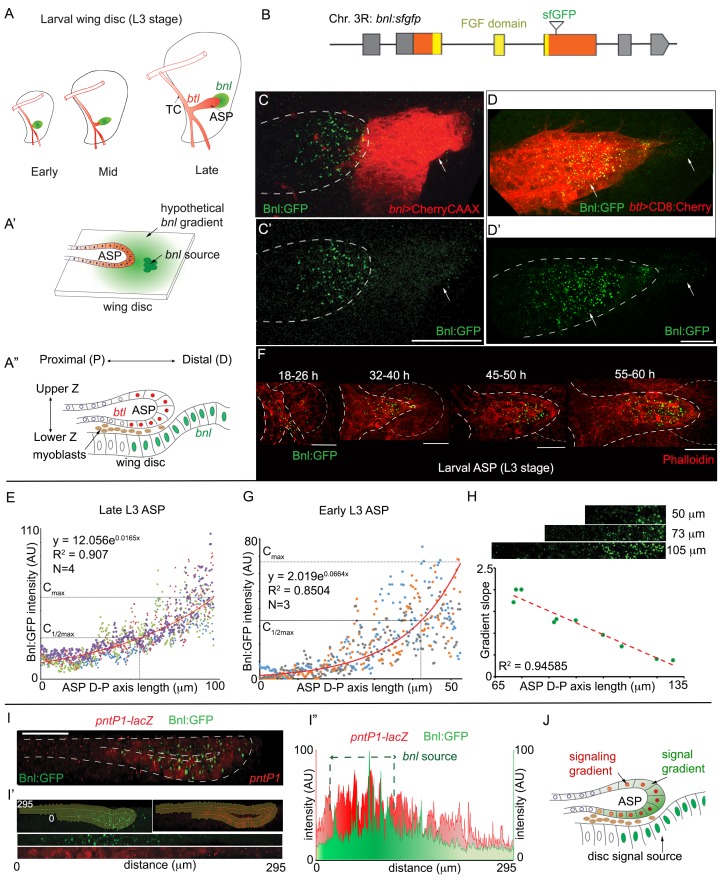
Figure 1. A concentration gradient of Bnl:GFP adopts precise morphologies of the recipient ASP.
(A) Drawings depicting budding and directed growth of the third instar larval ASP (red, btl expression) regulated by Bnl produced in a restricted group of cells in the wing disc (green); Bnl source spatiotemporally changes position ahead of the growing ASP; TC, transverse connective. (A’) Drawing depicting hypothetical chemotactic gradient of secreted Bnl (green) that was predicted to guide the directed ASP (red) growth toward the Bnl expressing cells (green circle). (A’’) Drawing depicting a cross-section of the late third instar larval ASP and wing disc, showing their epithelial contours, relative position in X-Z-Y dimension, putative Bnl- responsive cells (red), and disc bnl-expressing cells (green); upper/lower Z. (B) A schematic map of bnl:gfpendo knock-in allele; grey box, non-coding exons; orange box, coding exons; line, introns. (C–D’) Z-projected images showing that Bnl:GFP, produced at physiological level from the bnl:gfpendo allele, moved from the disc bnl-source to the ASP and distributed along the distal to proximal direction of the recipient tissue; (C,C’) bnl-source marked by CherryCAAX expression (bnl-LexA, lexO-CherryCAAX/bnl:gfpendo); (D,D’) recipient ASP marked by CD8:Cherry expression (btl-Gal4,UAS-CD8:Cherry/+; bnl:gfpendo); (C,D) merged red and green channels; (C’,D’) only the green channel; arrows, Bnl:GFP signal detected specifically in the wing disc source and recipient ASP. (E) Graph showing the Bnl:GFP concentration gradient along the ASP D-P axis in late third instar larvae (N = 4 independent samples). (F) Coordination of Bnl:GFP gradient formation with the ASP growth; time points, hours (h) after third instar larval molt; relative position of bnl-source marked by dashed-line. (G) Narrow range of Bnl:GFP gradient in shorter ASPs from early third instar larvae (N = 3 independent samples). (E,G) Red line graph, trend-line of the X-Y scatter plot with exponential fit from the averaged value; Cmax, maximum average Y value (Bnl:GFP intensity); (H) Negative correlation of the ASP D-P axis length and the slope (Cmax to C1/2max) of the Bnl:GFP gradient; each coordinate represents a single disc-ASP tissue; upper panels, Bnl:GFP distribution in three examples of ASPs with different lengths of D-P axis (the cropped region). (I) A 3D sagittal view showing a continuous long-range Bnl:GFP distribution across the entire recipient ASP epithelium adopting its tubular contour; expression of a target gene reporter of Bnl signaling, pntP1-lacZ (red, anti-βGal) showing corresponding signaling response; (I’,I’’) Intensity plots of Bnl:GFP (green) and pntP1-lacZ (red) (I’’) across the entire ASP epithelium derived from the digitally straightening ASP epithelium shown in I’. (J) Drawing of a cross section of the ASP-wing disc, summarizing the observations from C,D,I. (C–I’) Fixed samples, Z-projection, except I-I’; AU, arbitrary unit; dashed line, ASP or wing disc outline. Scale bars, 30 μm.