Figure 1.
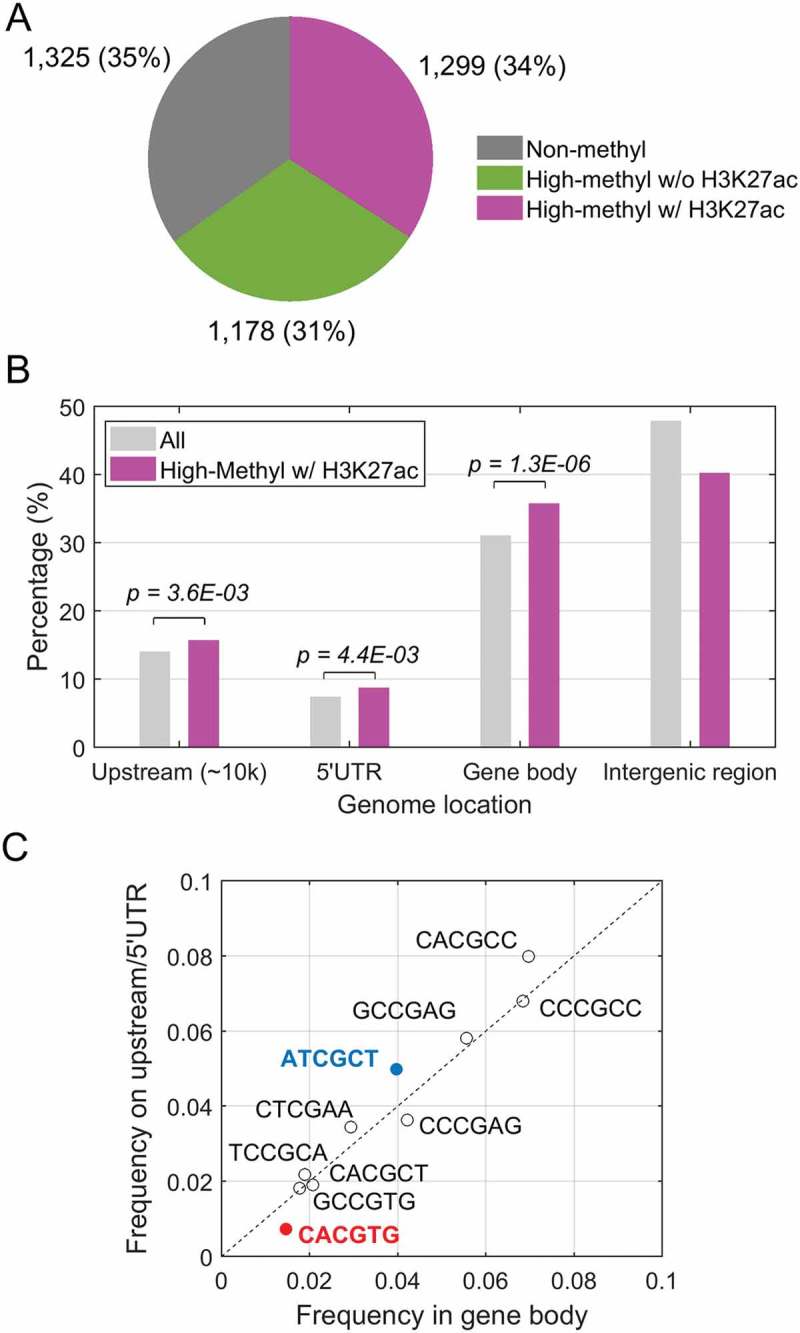
Global analysis of KLF4 binding to mCpGs at enhancer regions. A. Integration of KLF4 ChIP-seq, whole-genome bisulfite sequencing and H3K27ac ChIP-seq data in human glioblastoma U87 cells. A total of 1,299 highly methylated KLF4 binding sites were found to be occupied by enhancer marks as well. B. Distribution of the 1,299 KLF4 binding sites at 10 kb upstream, 5ʹUTR, gene body, and intergenic region. Compared with all KLF4 binding sites, KLF4-mCpG binding sites associated with the H3K27ac mark were significantly enriched in gene bodies. C. Motif analysis of the KLF4-mCpG binding sequences showing location preference at either gene body or upstream/5ʹUTR region.
