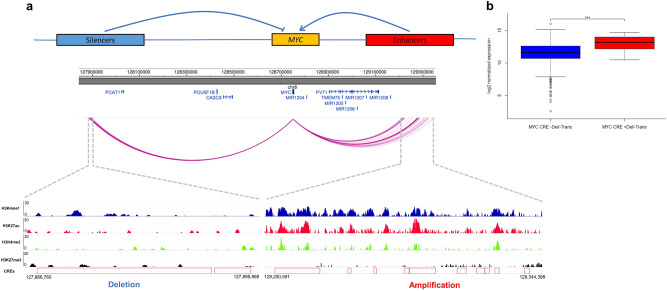
Fig. 3.
Copy number variations at cis-regulatory elements affect MYC gene expression in multiple myeloma. a Upper panel shows MYC gene expression may be regulated by CREs; CNVs at either the upstream putative silencers or downstream putative enhancers causing upregulation of MYC. Middle panel shows chromatin looping interactions between MYC promoter and CREs. Lower panel details ChIP-seq signals and relative positions of CNVs at these CREs in naïve B-cells. b CNV status at CREs and MYC expression. Difference in expression was assessed pairwise between samples with different CNVs status and the same translocation status by negative binomial test. ***P < 0.01. Trans translocation, Del deletion. From left to right, n = 345, n = 9, respectively. The hinges of the boxplot indicate the first and third quartile range