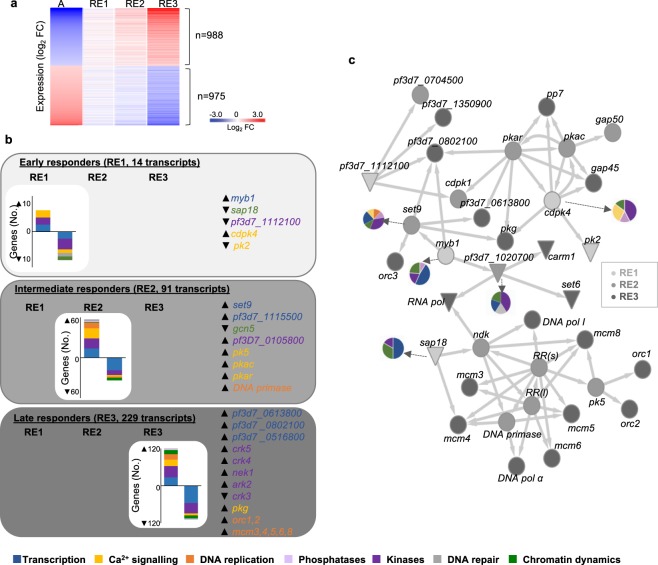
Figure 6.
Molecular mechanisms controlling cell cycle re-entry. The transcriptomes of parasites that re-entered their cell cycles (RE1–3) were analysed in context of genes matching key terms associated with cell cycle regulation using PlasmoDB (v33). (a) The expression profiles of genes of interest that were DE in RE1–3 are shown in heatmap compared to the arrested transcriptome (A = arrested). (b) The number of DE (either increased or decreased abundance) genes associated with specific cell cycle-related functional associations in histograms and genes of interest highlighted in grey boxes. Gene colours correspond to the GO functional classes as defined in the inserted key. (c) A gene association network was constructed between putative regulators of cell cycle re-entry by combining co-expression analysis (GRENITS, probability linkage score > 0.25) and functional association between genes (STRING v 10.0, combined probability score >0.8). Transcripts that were differentially expressed by RE1 (light grey), RE2 (grey) and RE3 (dark grey) are indicated according to their increased (●) or decreased (▼) transcript abundance. Genes of interest are indicated on the network, (RR = ribonucleoside-diphosphate reductase large (l) subunit pf3d7_1437200 and small subunit (s) pf3d7_1015800; orc3 = conserved unknown protein with orc3 domain, pf3d7_1029900) while the full gene association network is available in Supplementary File S4. Associated gene sets are indicated in inserted pie charts coloured according to GO annotations for gene functions.