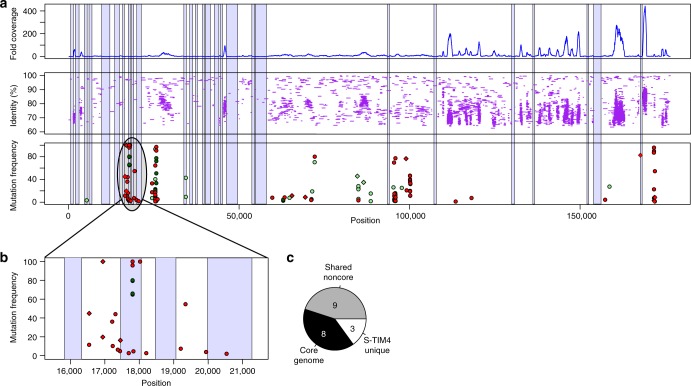
Fig. 5.
Variable genomic regions are not enriched in mutations. a Mapping of metagenomic reads to the S-TIM4 genome reveals mutations in variable and conserved regions with no enrichment in either. The position along the genome is on the X-axis. The lower panel shows mutation frequencies for phage populations along the genome. Colors denote the type of host the viral population evolved in: MIT9515 (dark green), MED4 (light green), and WH8102 (red). The middle panel shows the mapping of metagenomic reads to the S-TIM4 genome with the percent nucleotide identity of each read to the viral genome. The upper panel shows the fold-coverage of the metagenomic reads along the genome. Hypervariable genomic regions are shaded in purple (i.e. >500 bp long, coverage lower than 20% of mean). b Zoomed-in view of a mutation-rich genomic region (positions 15,500–21,500, open reading frames 23–27) of the S-TIM4 genome. c Adaptation of S-TIM4 to hosts occurs mostly by mutations in genes found in other cyanomyophages. Genes are classified as unique in S-TIM4 (have no close homologs in other phages), belonging to the cyanomyophage core genome (present in all studied cyanomyophage genomes), or shared noncore (appear in some cyanomyophage genomes). Raw data can be found in Supplementary Tables 2, 4