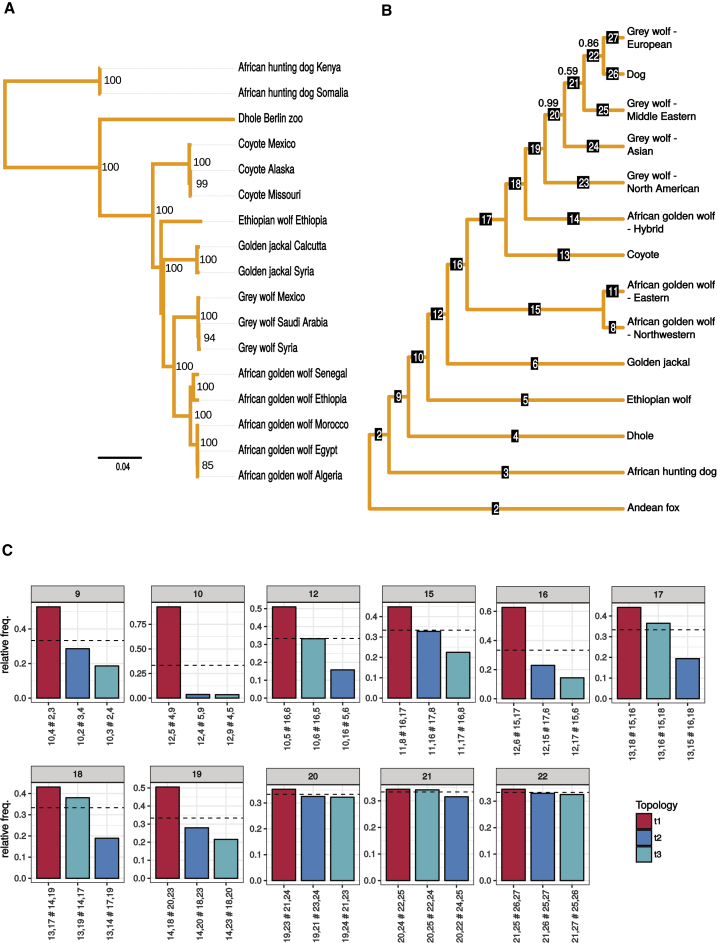
Figure 2.
Nuclear and Mitochondrial Phylogeny of Basal Canids
(A) The maximum-likelihood estimate of the mitochondrial phylogeny for a subset of the samples, using de novo mitochondrial assemblies obtained with MtArchitect. The node labels show the bootstrap support for the node.
(B) The phylogeny estimated from nuclear DNA by ASTRAL-II, where monophyletic clusters have been collapsed into a single leaf node. The tip labeled “African golden wolf-hybrid” represents a single known hybrid from the Sinai Peninsula—labeled “African golden wolf Egypt” in the mtDNA phylogeny—as described in the main text. The mean local posterior probabilities are shown for branches where this value is less than 1. The full nuclear phylogeny containing the sample relationships, branch supports, branch lengths proportional to divergence times, and estimated split times can be found in Figures S2A and S2B and Table S2.
(C) For a subset of the internal branches in the nuclear phylogeny, the quartet frequencies of the three possible configurations around each branch in the underlying unrooted tree are shown. The red bar represents the configuration shown in the phylogeny, and the two blue bars represent the two alternative configurations. For every quartet, the frequency of the true bipartition has previously been shown to be at least one-third [8], indicated here by a dotted line. Each alternative configuration is labeled by the bipartition it creates, with labels corresponding to those in (A). For example, the second bar of the panel labeled 12 swaps the positions of golden jackal (6) and Ethiopian wolf (5), whereas the third bar puts them as sister to each other. This plot summarizes the gene tree incongruence around examined branches.