Figure 1.
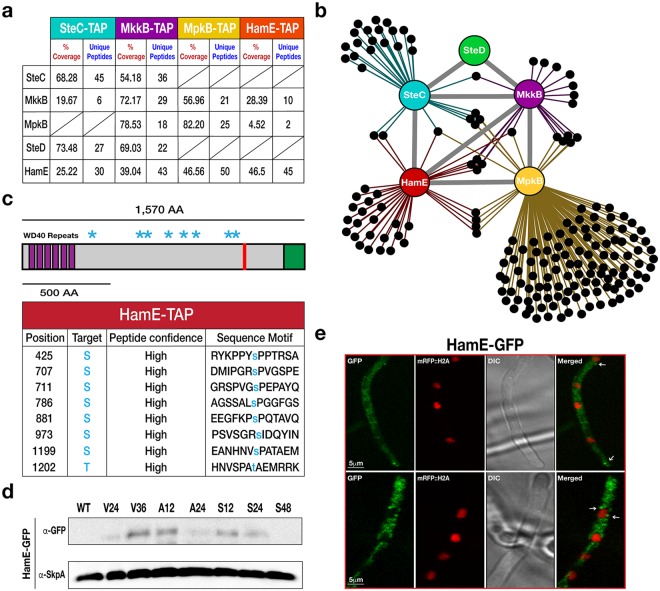
Discovery of the HamE scaffold protein interacting with components of the pheromone module in A. nidulans. (a) TAP pulldowns of the pheromone module kinases and HamE. TAP-tagged proteins are given at the top of the table and co-purified proteins are given on the left-hand side. The percentage of coverage and unique peptides of each detected protein are displayed. 2 biological replicates of each strain were used. (b) Interaction network of the pheromone module components based on unique peptides detected in each TAP pulldown. Each black dot represents a protein detected in two independent biological replicates but not in the wild type. (c) Schematic overview of the protein structure of HamE. HamE is a large, multi-domain protein that consists of 6 WD40 repeats at it’s N-terminus (aa residues 18–329). The red bar represents a coiled-coil domain (aa residues 1205–1225) and the green shaded area (1479–1570) represents a region of intrinsic protein disorder. Blue stars represent phosphorylation sites detected by mass spectrometry of TAP-tagged HamE. The amino acid positions and residues targeted for phosphorylation are listed in the accompanying table. S (serine), T (Threonine). (d) Time course immunoblotting of HamE at various stages of development. V (vegetative), A (asexual), S (sexual). For asexual and sexual induction, the HamE-GFP strain was cultured vegetatively for 24 hours in liquid GMM media and transferred to GMM plates to be incubated in the light and dark respectively. SkpA is used as a loading control. Full-length blots in (d) are presented in Supplementary Fig. 2. (e) Localisation of HamE-GFP in vivo at 16 hours of vegetative growth. The GFP fusion protein is dispersed throughout the cytoplasm and localises at the hyphal tips, cell membrane and nuclear envelope, indicated by white arrows.