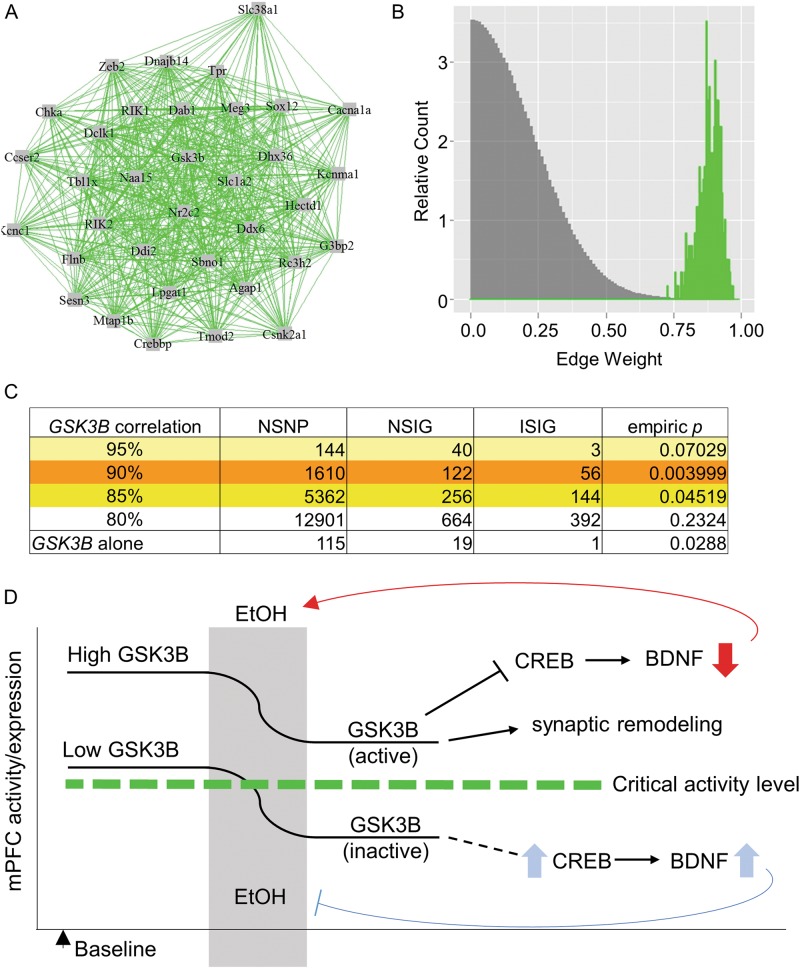
Fig. 5.
A Gsk3b-centered expression network in mouse mPFC is enriched for SNPs conferring increased risk for alcohol dependence in human GWAS data. a Network of genes with S-score, a measure of ethanol-responsiveness, correlation of r ≥ 0.9 with Gsk3b, found in the mPFC of BXD mice. b Comparative histogram of WGCNA-calculated edge weights (gene–gene expression correlations) of the 35 genes in the Gsk3b network (green) and edge weight values of 10,000 permutations of randomly selected groups of 35 genes (grey) from the BXD mPFC microarray. c SNP-set level analyses of human GWAS (COGA dataset, n = 1205 cases, 700 controls) were performed on groups of homologous genes. Results are shown for genes at multiple levels of correlation (80–95%) with Gsk3b in mouse mPFC. The result of a SNP-set analysis within the GSK3B gene alone is also given. NSNP = total number of genotyped SNPs in given gene or geneset; NSIG = number of significantly risk-conferring SNPs; ISIG = number of independent (non-LD) risk-conferring SNPs. Empiric p values were computed by 10,000 permutations of phenotypes. d Putative model by which mPFC GSK3B expression/activity may predispose to increased alcohol consumption and eventual dependence. Left side of diagram indicates a basal state prior to any ethanol exposure. We propose that high GSK3B activity is associated with high risk of increased ethanol consumption. GSK3B inhibition following ethanol exposure might normally serve to self-regulate ethanol consumption, such as through increased cortical BDNF which has been previously been shown to moderate drinking. With increased total GSK3B or increased basal GSK3B activity, there may be increased residual activity following a given ethanol exposure, allowing for continued GSK3B-mediated inhibition of BDNF, promoting the transition to increased consumption