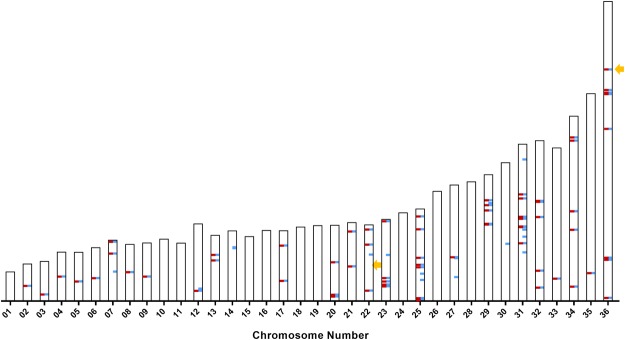
Figure 5.
Verification of previously identified SNPs and location of new SNPs that differ between the virulent VL and attenuated CL strains of L. donovani. Chromosomal location of previously identified homozygous non-synonymous SNPs between the cutaneous and visceral disease derived L. donovani strains (Red)4 compared to the novel SNPs identified only in this study (Blue) (synonymous and heterozygous codon changes identified are not labeled). Note that all the previously identified SNPs were also identified, or confirmed, in this study. 70 SNPs were previously identified across 66 genes. The same 70 SNPs were identified in this study, with an additional 15 novel SNPs not previously seen specific to the cutaneous strain. Genomic locations of SNPs identified in the previous study were translated to new genomic coordinates based on the new assembly for consistency. Arrows in yellow highlight the position of the previously identified RagC SNP on chromosome 36 and the A2 copy number difference on chromosome 22.