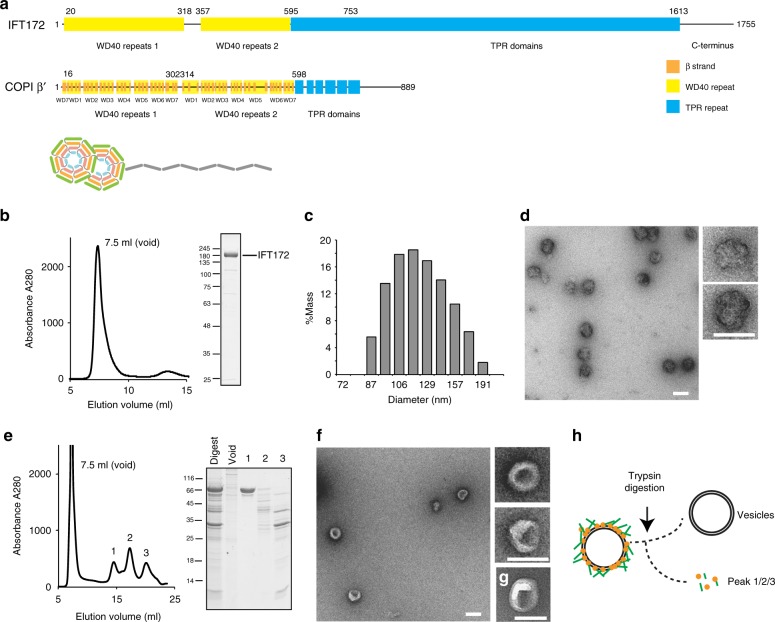
Fig. 1.
Characterization of recombinant Chlamydomonas IFT172. a Predicted domain architecture of IFT172 from Chlamydomonas. The predicted WD40 repeats (in yellow) and TPR (tetratricopeptide repeat) domain (in blue) were defined by HHpred. IFT172 shows a similar domain organization to COPI/II and clathrin. COPI β’ from Saccharomyces cerevisiae is shown for comparison. b Size-exclusion chromatography (SEC) profile of recombinant full-length IFT172 and Coomassie-stained SDS-PAGE analysis of recombinant IFT172 after size-exclusion chromatography. c Dynamic light scattering (DLS) measurement of IFT172 shows a uniform particle size distribution of IFT172 with an average diameter of ~120 nm with 100% mass. d Negative stain EM micrograph of IFT172 showing globular oligomer formation. Scale bar: 100 nm. e SEC profile of IFT172 proteolyzed products after limited-proteolysis (left) and SDS-PAGE of the peak fractions from SEC (right). f Negative stain EM micrograph of the fraction from the void peak in e shows round particles with smooth surface. Scale bar: 100 nm. g Negative stain EM micrograph of the liposomes from Folch fraction I as a control. Scale bar: 100 nm. h Scheme describing the experimental procedure of e and b