Fig. 2.
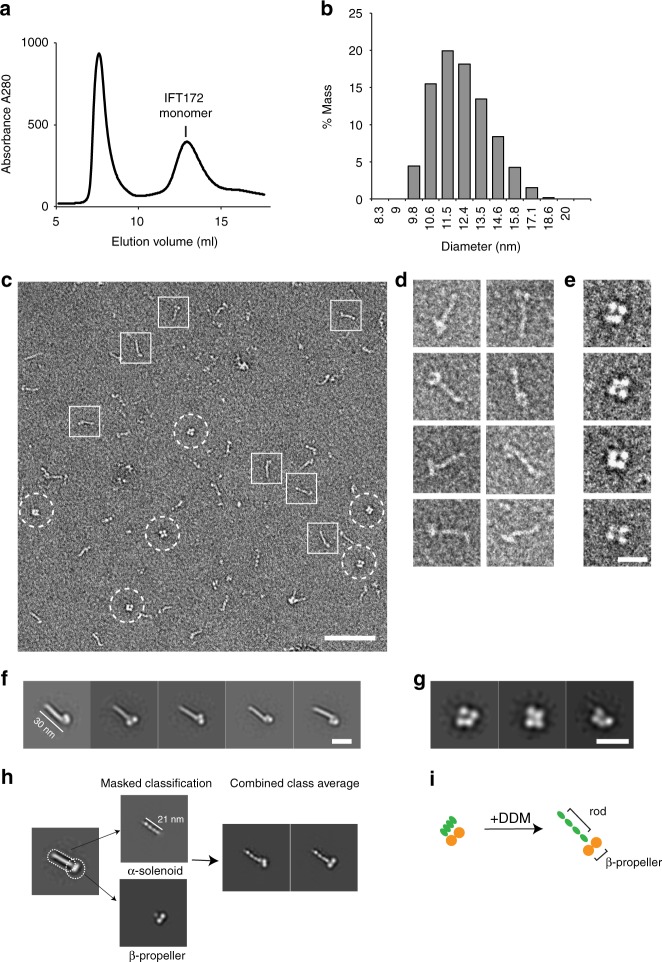
Monomeric architecture of recombinant Chlamydomonas IFT172. a SEC profile of recombinant full-length IFT172 with 0.1% DDM shows an increased amount of the monomer peak. b DLS measurement of IFT172 monomer shows the size of the particle in the monomer peak is around 11 nm. c Negative stain EM micrograph of monomeric IFT172 with 0.1% DDM, showing an open (square) and a closed (circle) conformation. Scale bar: 100 nm. d, e Representative IFT172 monomers in an open (d) and closed (e) conformation. Scale bar: 20 nm. f, g. Reference-free 2D class averages of IFT172 in an open (f) and a closed (g) conformation. Scale bar: 20 nm. The open conformation shows a rod-like structure with 30 nm in length, while the closed IFT172 contains 4 sub-domains. h Detailed 2D averages of IFT172 by domain masking and assembly strategy. Scale bar: 20 nm. The original average views are shown on the left side. To obtain detailed features for each domain, soft masks (dotted lines) were applied to facilitate an individual alignment of the head domain and tail domain. Two complete IFT172 molecular are generated by aligning the two components. The N-terminal head region shows two globular densities with a size of ~5 nm each. The rod part shows 4 globular domains with 4 nm each, which could be the features by TPR alpha solenoid motifs. i Schematic representation of the conformational change of monomeric IFT172 induced by DDM