Figure 4.
Two Layers of Negative Regulatory Loops Determine MITF Damped Oscillatory Behavior
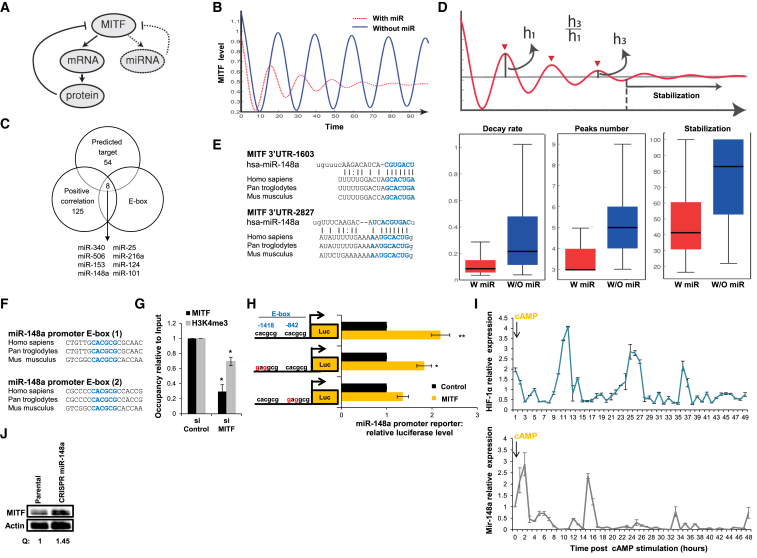
(A) Mathematical model depicting MITF negative regulation by PDE4D and HIF1α (bold, solid lines indicate that transcription and translation are required) and miRNAs (dashed lines indicate that only transcription is required).
(B) A simulation of MITF expression dynamics as derived from the mathematical model under negative regulation of protein only (blue) or a combination of protein and miRNA (red).
(C) Overlap between 62 miRNAs predicted to target MITF (http://www.targetscan.org), 133 miRNAs in positive correlation with MITF (Bell et al., 2014), and miRNAs that include E-box in their promoter.
(D) Upper panel: an illustration depicting the dynamical features used for comparison between the reference (protein-derived regulation) and the reference under an additional miRNA regulation. Lower panels indicate oscillation features: decay rate (corresponding to the ratio between the third and first peaks, left panel), number of peaks (middle panel), and time to reach stabilization (right panel) under negative regulation of either protein (blue) or a combination of protein and miRNA (red).
(E) Predicted binding site for miR-148a in the MITF 3′UTR sequence.
(F) Two conserved MITF DNA binding sites (E-boxes) in miR-148a promoter sequence.
(G) ChIP was performed on extracts from WM3682 cells transfected with siMITF or siControl. Protein:chromatin-crosslinked complexes were precipitated with the indicated antibodies. PCR primers spanning the region encoding the miR-148a promoter were used. The data show promoter occupancy relative to input. Error bars represent SEM. ∗p < 0.05 (n = 3).
(H) Expression of miR-148a promoter reporter with WT or mutated E-box regions upon MITF cDNA (MITF) or empty vector (control) expression in HEK293T cells. Firefly luciferase activity was normalized to Renilla luciferase activity. Fold changes relative to control are indicated. Error bars represent SEM. ∗p < 0.05; ∗∗p < 0.01 (n = 3).
(I) HIF1α and miR-148a levels were quantified in MNT-1 samples collected hourly for 48 hr upon cAMP stimulation. Levels were normalized to actin and U6, respectively. Error bars represent SEM of technical replicates (n = 3).
(J) MITF protein levels in parental WT MNT-1 cells (control) and the miR-148a-deleted MNT1 cell line. Actin was used as a loading control. Quantification of protein amount normalized to actin (Q) is indicated.