Figure 6.
A Complex of ROD-1 without β-Propeller and CZW-1Zw10 Self-Assembles Efficiently into Higher Order Oligomers In Vitro
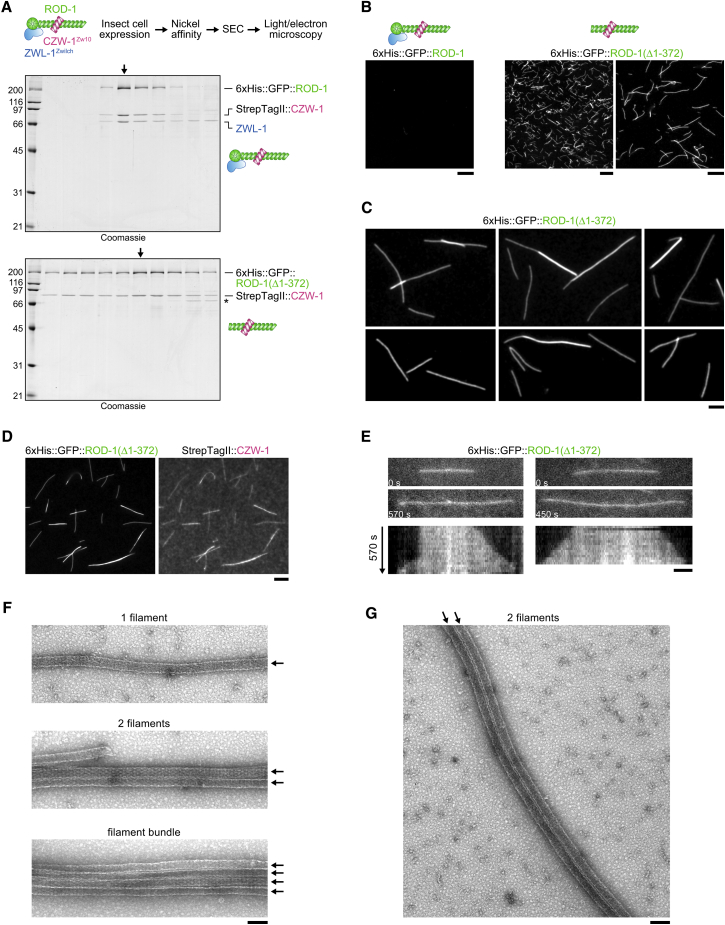
(A) (Top) Workflow used to generate the in vitro data shown in this figure. (Bottom) Coomassie-stained gels showing the protein fractions after SEC. Fractions analyzed in subsequent panels are marked with an arrow. Note that purity and amount are comparable for both complexes. Molecular weight is indicated in kDa on the left.
(B) Images of the protein fractions marked with an arrow in (A) examined by fluorescence light microscopy, showing that GFP::ROD-1(Δ1-372), but not full-length GFP::ROD-1, oligomerizes into micrometer-long filaments with high efficiency. Scale bars, 10 μm for left and middle image; 5 μm for right image.
(C) Higher magnification views of GFP::ROD-1(Δ1-372) filaments showing evidence of lateral bundling. Note that filaments reach up to 15 μm in length. Scale bar, 2 μm.
(D) Fluorescence image confirming that GFP::ROD-1(Δ1-372) co-localizes with StrepTagII::CZW-1Zw10 on filaments in vitro. Scale bar, 5 μm.
(E) Selected images (top) and corresponding kymographs (bottom) from a time-lapse sequence (30 s between frames), showing that GFP::ROD-1(Δ1-372) filaments grow from both ends. In this experiment, filaments were directly examined after the nickel affinity step. Scale bar, 2 μm.
(F and G) Transmission electron microscopy images of GFP::ROD-1(Δ1-372) filaments. Individual filaments (arrows) have an invariant diameter of ∼50 nm and tend to associate with each other laterally (F), often over distances of several micrometers (G). Scale bars, 100 nm.
See also Figure S6.