Figure 5.
EJC Depletion Leads to Inclusion of Cryptic Microexons
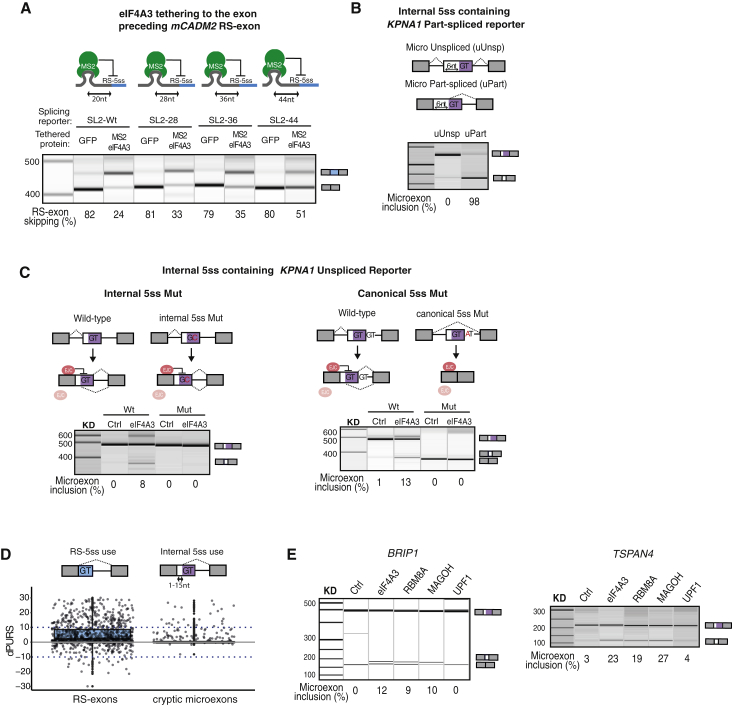
(A) The SL2 in mCADM2 splicing reporter was moved upstream of the exon-exon junction as indicated, and the RS-exon splicing pattern was analyzed after co-transfection of SL2 reporters and plasmids expressing MS2-eIF4A3 or GFP proteins.
(B) Six nucleotides (GCACAG) were added at the beginning of the KPNA1 RS-exon to move the RS site to an internal 5ss, thus creating a KPNA1 microexon reporter. Unspliced and part-spliced version of the reporters were transfected, and their splicing pattern was analyzed by RT-PCR.
(C) The internal or canonical 5ss within KPNA1 microexon unspliced reporter were mutated, and the splicing pattern was analyzed by RT-PCR after transfection into HeLa cells treated with control or eIF4A3 siRNAs.
(D) The difference in the use of RS-5ss or internal 5ss in EJC KD compared to control (dPURS) is shown in HeLa cells. Cryptic microexons result from the use of an internal 5ss that is located within the first 15 nt of a longer annotated exon. Positive dPURS values indicate increased RS-exon skipping or increased inclusion of cryptic microexons.
(E) RT-PCR validation of 2 microexon inclusion events identified in (D) after KD of EJC core factors or UPF1.
Results shown in (A)–(C) and (E) derive from a minimum of 3 independent replicates performed in HeLa cells.