Figure 6.
EJC-Mediated Repression of Recursive Splicing in the Brain, and Physiologic Recursive Splicing
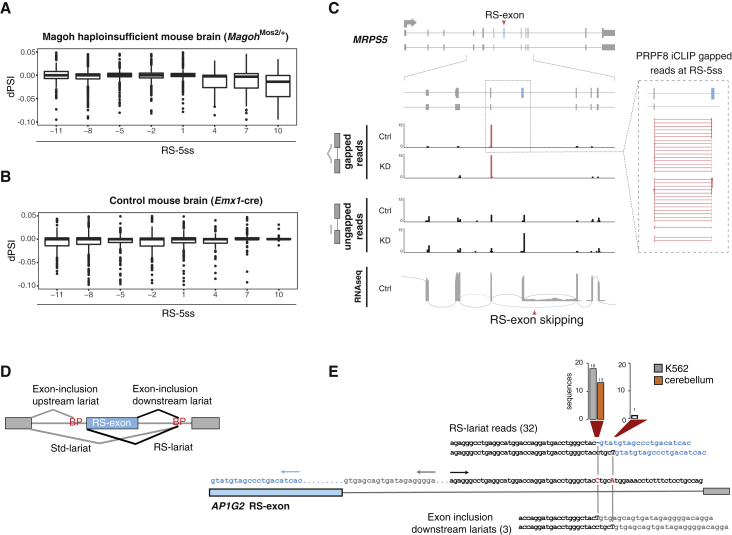
(A) The dPSI of exons highly included in wild-type mouse brain (PSI > 90%) is shown after comparison with Magoh haploinsufficient mouse brain as in Figure 1E.
(B) Same as (A), but comparing control Emx1-cre and wild-type mouse brain.
(C) A schematic of the MRPS5 gene with the RS-exon highlighted in blue. Below, PRPF8 iCLIP crosslinking is shown, as identified by either ungapped or gapped reads in control or eIF4A3 KD cells (4 replicates are summed up per condition). Crosslinks upstream of junctions involving the RS-exon are shown in red, and corresponding reads are zoomed into on the right. Further below, a Sashimi plot shows RNA-seq evidence of RS-exon skipping.
(D) The types of possible lariats associated with RS-exon splicing are named. In black are the lariats that were detected for the AP1G2 gene as described in (E).
(E) Sequence of the RS-exon in the AP1G2 gene and its downstream intron. BP annotations based on lariat sequencing are highlighted in red. Arrows indicate primers used to interrogate lariats shown in black in (D). Above, sequences and number of reads supporting RS-lariats (inverted alignment) with alternative BP annotations in cerebellum and K562 cells are shown. Below, sequences and number of reads supporting exon inclusion downstream lariats with alternative BP annotations are shown.