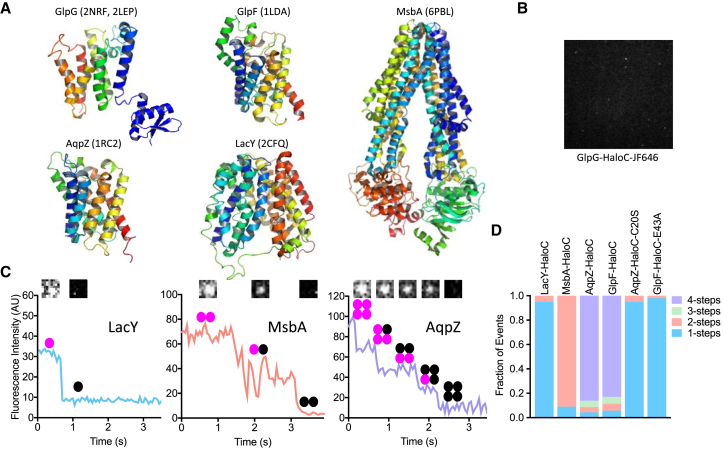
Figure 2.
Quantitative photobleaching of single molecules of E. coli inner membrane proteins. (A) X-ray crystal structures depicting lateral/membrane views (periplasm up, cytosol down) of E. coli rhomboid GlpG and four other E. coli inner membrane proteins that were used as oligomer standards. Proteins are colored from blue (amino terminus) to red (carboxy terminus), and Protein Data Bank (PDB) accession numbers used to render images are included in the brackets. Note that structures of the GlpG membrane core and cytoplasmic domain that were crystallized separately are represented together in arbitrary conformation relative to each other. Minimal functional units are monomers for all except MsbA, which is dimeric, as shown. (B) A representative single-molecule TIRF microscopy image of GlpG-HaloC in a planar supported lipid bilayer. (C) Representative fluorescence intensity versus time traces of LacY-HaloC, MsbA-HaloC, and AqpZ-HaloC single molecules labeled with HTL-JF646. Insets show corresponding images and oligomer diagrams of the single-molecule spots before and after each photobleaching step. Note the distinctive steps of intensity drops as the fluorophores photobleach. (D) Quantification of photobleaching steps of bacterial proteins in supported lipid bilayers. Rare events (e.g., two-step photobleaching events in monomeric samples) are expected because molecules can be unassociated but co-occupy a space smaller than the resolution of a fluorescence microscope. To see this figure in color, go online.