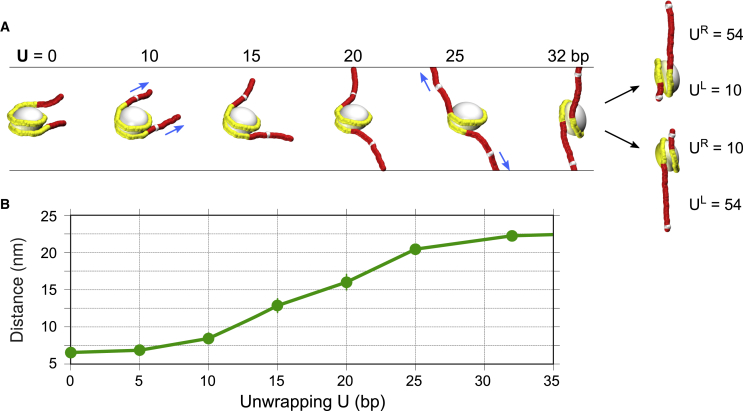
Figure 4.
Gradual unwrapping of nucleosomes facilitates fiber stretching. (A) Typical conformations of nucleosomes in extended fibers. For unwrapping U = 0, all nucleosomal DNA is attached to histones (shown in yellow). For U > 0, the unwrapped DNA is represented by the red fragments located between the ends of the yellow fragments and the white markers (NCP positions 1 and 145). An increase in U brings about an increase in the angle between the DNA end vectors (arrows starting at the white markers) so that for U = 25–32 bp, this angle is ∼180°, with the end vectors perfectly aligned along the external force. The alternative asymmetric conformations on the right, with UL and UR equal to 10 or 54 bp, have the same average degree of unwrapping as the U = 32 bp case and generate the same level of extension (Zext). However, the form of the adhesion energy profile and the stochastic dynamic of unwrapping-rewrapping dictates that these asymmetric conformations are preferable (Figs. 5 and S8). (B) The nucleosome end-to-end distance (between the white markers in (A)) as a function of unwrapping, U. The largest increase in this distance occurs between U = 10 and 25 bp, which roughly corresponds to the plateau limits (Fig. 2). To see this figure in color, go online.