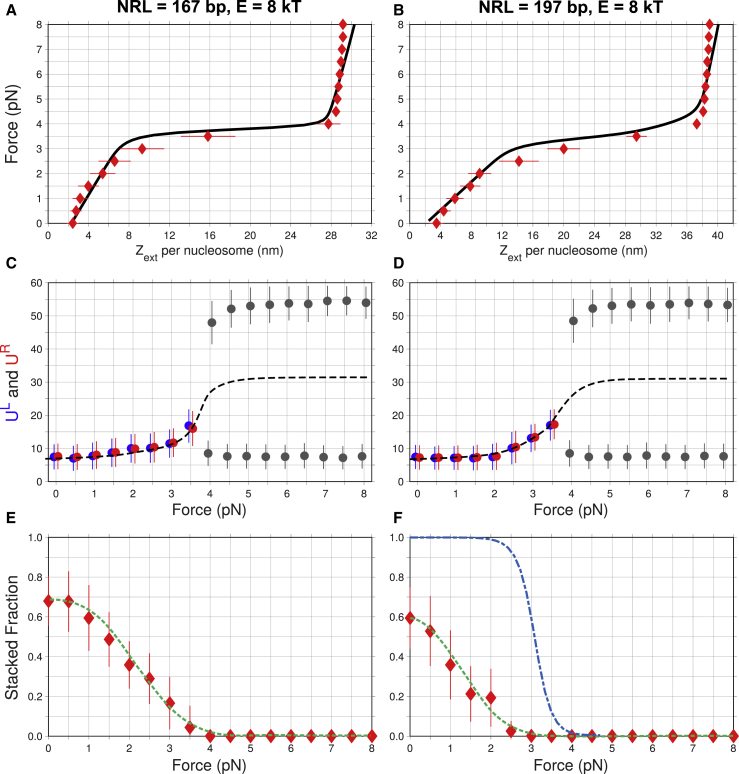
Figure 5.
Response of the nucleosomal arrays (extension, unwrapping, and stacked fraction (SF)) to the external force. (A and B) The MC force-extension profiles (red diamonds) were calculated using the adhesion potential, Gadh(U), for DNA unwrapping (Fig. S2) and E = 8 kT for stacking interactions. Note that the linear response persists up to F = 2.0–2.5 pN in both cases. The solid curves represent the adjusted experimental data (15). (C and D) DNA unwrapping at the left and right ends of nucleosomes (UL in blue and UR in red) is shown as a function of force. The average values are shown as circles, and SDs are shown as bars. As the external force increases from 0 to 3 pN, the DNA unwrapping (UL and UR) gradually changes from 8 to 12 bp, with an SD ∼4 bp. These values are the averages over all 12 nucleosomes in the array and are in accord with our findings for the uniform U values (Figs. 2 and S7). At F ≥ 4.0 pN, the DNA unwrapping undergoes bifurcation into two branches with U ≈ 8 and 54 bp. Here, the circles and bars are shown in gray (i.e., not in red or blue), because asymmetric unwrapping occurs stochastically, that is, on average the chances of each nucleosome end (UL or UR) to be strongly unwrapped are equal (see Figs. S2 and S8 for details). The dashed curves represent the average unwrapping for both nucleosomal ends and exhibit a sigmoidal trend. (E and F) Average SFs of nucleosomes in fibers are shown as a function of force. Stacking energy is E = 8 kT. The sigmoidal blue curve is a result of statistical-mechanical modeling by Meng et al. (Fig. 3 D in (15)). The dashed green curves are sigmoidal fits over the MC results. Note that we do not observe a fully stacked fiber (SF = 1) at any force. To see this figure in color, go online.