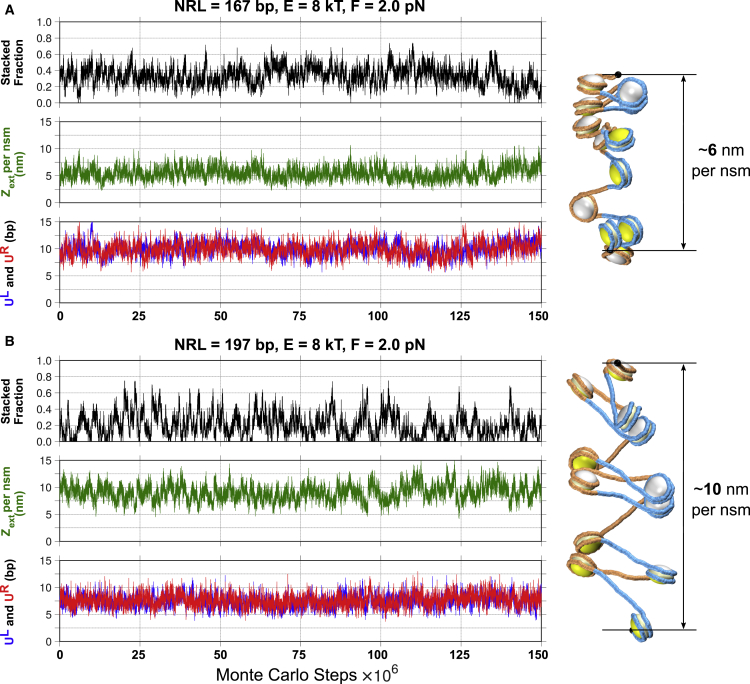
Figure 6.
Changes in the fiber parameters during MC simulations show dynamic unwrapping and stacking of fibers. 150 × 106 MC equilibrated steps are shown for NRL = 167 bp (A) and 197 bp (B) at F = 2 pN. (Top) The fraction of stacked nucleosomes is shown. Equilibrium involves multiple stacking-unstacking transitions. (Middle) The extension per nucleosome is presented. The SD of extension is ∼1.1 nm for NRL = 167 bp construct and ∼1.4 nm for 197 bp. (Bottom) UL and UR are shown as a function of MC steps. Note that these UL and UR values are the averages over 12 nucleosomes in the array. Examples of the equilibrated conformations are shown on the right. Note that the structures became half open during simulations. They show the ∼6-nm vs. ∼10-nm extensions consistent with the experimental data (15); see Fig. 5, A and B. To see this figure in color, go online.