Table 4.
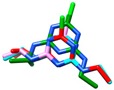
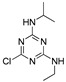
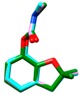
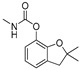
Training set pesticides of the highest toxicity with the unknown crystal structures: chemical structures, conformational analysis, superposition of the experimental conformation and generated global minima using the best performing force fields.
| Pesticide | FF a | Eglob_min b | NGMS c | NF d | Pesticide | FFDAA f |
|---|---|---|---|---|---|---|
| (kJ/mol) | Alignment e | RMSD (Å) g | ||||
| atrazine | MM3 | −1161.48 | 445 | 23 |
|
MM3/MMFF |
|
AMBER94 | NA h | NA | NA | 0.892 | |
| MMFF | −1007.32 | 147 | 62 | MM3/MMFFs | ||
| MMFFs | −992.63 | 509 | 20 | 0.931 | ||
| OPLSAA | −783.57 | 364 | 19 | MMFF/MMFFS | ||
| 0.343 | ||||||
| carbofuran | MM3 | 29.60 | 3617 | 2 |
|
MMFF/MMFFS |
|
AMBER94 | NA | NA | NA | 0.075 | |
| MMFF | 20.30 | 3307 | 2 | MMFF/OPLSAA | ||
| MMFFs | 15.92 | 3384 | 2 | 0.216 | ||
| OPLSAA | 73.27 | 5621 | 75 | MMFFs/OPLSAA | ||
| 0.239 |
a Force field; b Energy of the global minimum; c Number of times that a single global minimum structure was found in 10,000 processed structures; d Number of families i.e., different conformations found in 10,000 processed structures; e Red—experimental conformation, Violet—MMFF conformation, Blue—MMFFs conformation, Green—OPLSAA conformation; f Force field dependent alignment accuracy; g RMSD measured between the heavy atoms of pesticides experimental and best performing force field conformations; h Not available.
