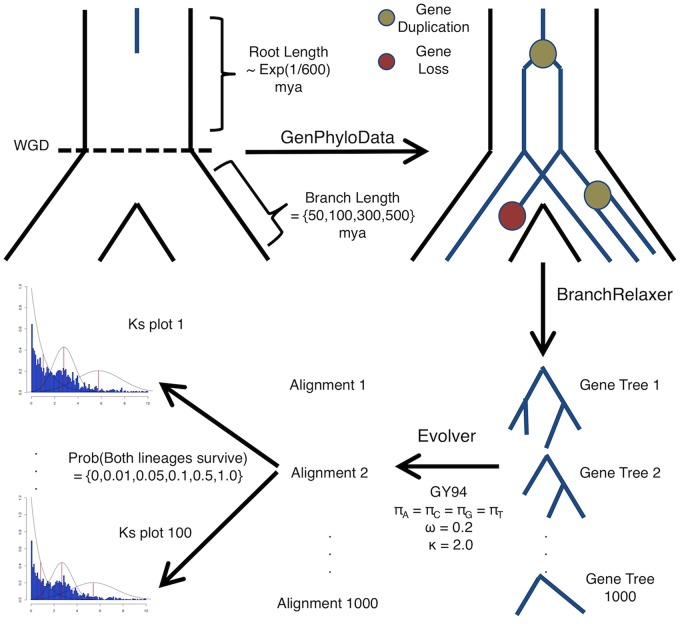
Fig. 1.
—Stylized workflow for simulating gene trees with a known WGD under a known retention rate. Gene families with an explicit WGD are simulated similar to gene trees evolving within a species tree. The branch length of the root of the species tree is drawn from a distribution so not all gene families are single copy before the WGD. The branch lengths following the WGD either are 15, 50, 100, 200, 300, or 500 Ma, which correspond to Ks of 0.15, 0.5, 1.0, 2.0, 3.0, and 5.0, respectively. Gene trees then evolve under variable rates of gene duplication and loss, followed by relaxation of branches to account for substitution rate variation. Sequence data was then simulated on these relaxed gene trees. We implemented a nonstandard data resampling procedure here that allowed for random gene loss. Individual gene trees were not resampled, but the probability that both lineages following a WGD survived was resampled. One hundred Ks plots were then generated from the 100 resampled data sets of 1000 trees.