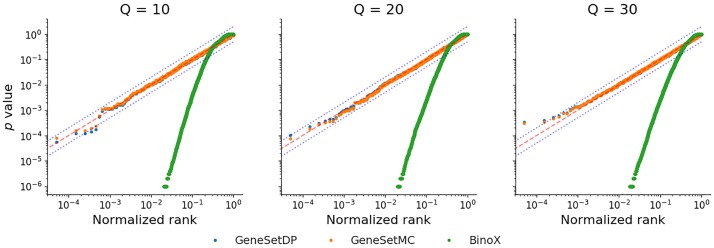
Fig 2. Calibration of GeneSetDP and GeneSetMC.
We selected 10000 random sets of 10, 20 and 30 genes from the investigated genome definition and calculated their p values with GeneSetMC and GeneSetDP against the “Glycolysis/Gluconeogenesis” pathway. To test the p values uniformity we plotted them against their normalized rank, i.e. each p value’s (<rank>-0.5)/<total number of p values>. We also added the calibration curve of BinoX, evaluated on the same random sets. The dashed line shows y = x and the dotted lines y = 2x and y = 0.5x, for comparison.