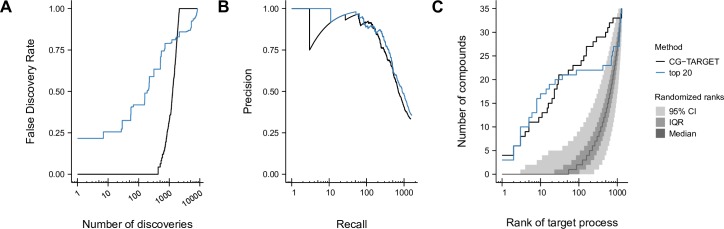
Fig 3. Comparison of CG-TARGET performance versus gene-target enrichment.
Perturbed bioprocesses were predicted using both CG-TARGET and a method that calculated enrichment on the set of each compound’s 20 most similar genetic interaction profiles (“top 20”). (A) Bioprocess prediction false discovery rate estimates derived from resampled chemical-genetic interaction profiles, performed on compounds from the RIKEN dataset. (B) Precision-recall analysis of the ability to recapitulate gold-standard annotations within the set of top bioprocess predictions for ~4500 simulated compounds. Each simulated compound was designed to target one query gene in the genetic interaction network and thus inherited gold-standard biological process annotations from its target gene. (C) For each of 35 well-characterized compounds in the RIKEN dataset with literature-derived, gold-standard biological process annotations, we determined the rank of its gold-standard bioprocess within its list of predictions. The number of compounds for which a given rank (or better) was achieved is plotted. The grey ribbons represent the median, interquartile range (25th to 75th percentiles), and 95% confidence interval of 10,000 rank permutations.