Abstract
Lipopolysaccharide (LPS) elicitors isolated from Pseudomonas fluorescens UOM SAR 14 effectively induced systemic and durable resistance against pearl millet downy mildew disease caused by the oomycete Sclerospora graminicola. Rapid and increased callose deposition and H2O2 accumulation were evidenced in downy mildew susceptible seeds pre-treated with LPS (SLPS) in comparison with the control seedlings, which also correlated with expression of various other defense responses. Biochemical analysis of enzymes and quantitative real-time polymerase chain reaction data suggested that LPS protects pearl millet against downy mildew through the activation of plant defense mechanisms such as generation of nitric oxide (NO), increased expression, and activities of defense enzymes and proteins. Elevation of NO concentrations was shown to be essential for LPS-mediated defense manifestation in pearl millet and had an impact on the other downstream defense responses like enhanced activation of enzymes and pathogen-related (PR) proteins. Temporal expression analysis of defense enzymes and PR-proteins in SLPS seedlings challenged with the downy mildew pathogen revealed that the activity and expression of peroxidase, phenylalanine ammonia lyase, and the PR-proteins (PR-1 and PR-5) were significantly enhanced compared to untreated control. Higher gene expression and protein activities of hydroxyproline-rich glycoproteins (HRGPs) were observed in SLPS seedlings which were similar to that of the resistant check. Collectively, our results suggest that, in pearl millet-downy mildew interaction, LPS pre-treatment affects defense signaling through the central regulator NO which triggers the activities of PAL, POX, PR-1, PR-5, and HRGPs.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1501-y) contains supplementary material, which is available to authorized users.
Keywords: Pearl millet downy mildew, Lipopolysaccharide, Induced resistance, Nitric oxide, Defense enzymes, PR-proteins gene expression
Introduction
Pearl millet [Pennisetum glaucum (L.) R. Br.] being more tolerant to drought stress serves as an important cereal crop, both in terms of food and fodder for millions of people living in the semi-arid regions of Asia and Africa. As a staple food, nearly, about 50 million Indians, particularly in the states of Rajasthan, Maharashtra, and Gujarat are dependent on pearl millet and it is grown in an area of seven million hectares with an annual production of 9.25 million tonnes (Yadav 2014; Prakash et al. 2014). Downy mildew is the most destructive disease of pearl millet caused by the biotrophic oomycete Sclerospora graminicola (Sacc.) Schroet severely affecting the production thereby incurring huge economic burden to the farmers (Thakur et al. 2011; Nayaka et al. 2017). Popular and conventional methods like resistance breeding, chemical control, and biological control are employed for disease management; however, each method has several limitations. Chemical control of downy mildew is uneconomical and associated with safety risks; resistant hybrids succumb to the breakdown of resistance due to the highly variable nature of the pathogen. Therefore, it is essential to explore eco-friendly, economically feasible, and safer alternatives for disease management.
The resistance against pathogens can be triggered in plants through a phenomenon termed induced resistance which is an eco-friendly and safer alternative strategy for plant protection. Several studies across different plant–pathogen systems have demonstrated the efficacy of induced resistance by minimizing the use of hazardous chemicals. Induced resistance can be achieved by an array of biotic and abiotic agents which act by eliciting an increased expression of innate immunity of plants against a wide range of pathogens (Lyon 2007; Walters et al. 2013). Among the several determinants involved in the induction of systemic resistance by plant growth promoting rhizobacteria (PGPR), the important are lipopolysaccharides (LPS) present in the outer membrane of gram-negative bacterial cells (Ramamoorthy et al. 2001; Van Loon et al. 1998). LPS is a ubiquitous and indispensable component of the membrane of gram-negative bacteria which recognized by plants to trigger some plant defense-related responses (Dow et al. 2000; Erbs and Newman 2012; Newman et al. 2007). Coventry and Dubery demonstrated that the pre-treatment of tobacco plants (Nicotianae tabacum) with LPS isolated from a bacterial endophyte Burkholderia cepacia, protected tobacco plants from Black-shank disease caused by Phytophthora nicotianae. The LPS treatment was shown to enhance the activity of several PR-proteins (Coventry and Dubery 2001; Uzma et al. 2018). LPS derived from Enterobacter asburiae induced systemic resistance in lettuce against soft rot disease caused by Pectobacterium catovorum subsp. catovorum (Pcc), and this was associated with increased activities of the defense enzymes such as superoxide dismutase and peroxidase (Jetiyanon and Plianbangchang 2013).
Induction of systemic and durable resistance against downy mildew of pearl millet has been demonstrated in earlier studies from our laboratory using LPS from Pseudomonas fluorescens isolate UOM SAR 14 (Niranjan-Raj et al. 2011). However, the mechanism of resistance was not studied in our previous investigation. In the present study, we investigated the mechanism of resistance by activity and expression of defense enzymes in downy mildew of pearl millet on induction with LPS.
Experimental procedures
Host
Two pearl millet cultivars were used as host plants for all the experiments. The downy mildew susceptible plants were raised from the seeds of the pearl millet cv. 7042S and the resistant plants were raised from the seeds of the pearl millet cv. 18292. Both 7042S and 18292 were sourced from International Crop Research Institute for Semi-Arid Tropics (ICRISAT), Hyderabad, India and All India Coordinated Research Project on Pearl Millet (AICRP-PM), Mandore, Rajasthan, India.
Pathogen source and preparation of inoculum
The downy mildew susceptible 7042S pearl millet crop was raised in the downy mildew sick plot which is heavily infested with Sclerospora graminicola oospores. The downy mildew pathogen was maintained throughout the experimental period on this susceptible cultivar under field conditions. The 7042S seeds were coated with the downy mildew oospores and were hand-sown in the downy mildew sick plot which provided additional inoculum in the form of soil oospores. The seedlings which emerged showed downy mildew infection from the two-leaf stage onwards and produced sporangia on their abaxial side. Such infected seedlings were maintained by providing necessary irrigation and fertilization. The downy mildew zoospores collected from the affected leaves were used as inoculum. In the evening hours, diseased leaves with high sporulation on the abaxial side were collected, washed thoroughly in running tap water until the sporangia are removed. These leaves were cut into small pieces, blotted dry and were arranged in Perspex plates (lined with moist blotters on both sides) and were kept in a dark, moist chamber for sporulation. The following morning, the leaf pieces were observed for sporulation and the fresh crop of sporangia was scraped into the sterile distilled water. The final concentration of the zoospores was adjusted to 40,000/ml using a hemocytometer and used for inoculation.
LPS preparation and seed treatment
The bacterial isolate P. fluorescens UOM SAR 14 was used as the source for the extraction of LPS. The extraction, preparation, and pearl millet seed treatment of LPS were carried out as described previously (Niranjan-Raj et al. 2011). Pseudomonas fluorescens UOM SAR 14 was inoculated on to King’s medium base agar (27 °C) and incubated for 24 h. The bacterial cells were harvested, washed using phosphate buffered saline (10 mM, pH 7.2) by centrifugation at 4000 g for 5 min, followed by lyophilization. The cells were further suspended in 50 mM Tris–HCl and 2 mM EDTA buffer (pH 8.5) and subsequently sonicated at resonance amplitude for six times for 30 s at 0–4 °C. The intact cells were separated by centrifugation, and the supernatant was further centrifuged at 8000 g for 60 min to obtain a pellet which was resuspended in 2 mM Tris–HCl buffer of pH 7.8 and stored at − 20 °C. Thereafter, the pellet suspension which contains crude LPS, proteins, and nucleic acids was repeatedly dialyzed against water. Furthermore, the pellet was lyophilized and was extracted with phenol/chloroform twice. The resulting aqueous phase was pooled, again lyophilized, and suspended in Tris–HCl buffer. The water phase was vigorously shaken and centrifuged at 16,000 g for 5 min to separate and collect the LPS. The water phase was shaken with an equal volume of chloroform to further remove the traces of phenol. Protein, DNA, and RNA contamination were removed by treating the aqueous phase Proteinase K, DNase, and RNase, respectively. The resultant aqueous phase was vacuum dried, resuspended in Tris–HCl buffer, and stored at -20 °C. The purity of the LPS preparation was checked by mixing LPS sample with 1 µl of bromophenol blue, boiled and cooled for a minute and centrifuged at a speed of 12,000×g. This sample was subjected to SDS-PAGE on 10% gels run at constant voltage (90V) and varying current (≤ 40 mA) at 4 °C. The gels were stained using the silver stain and scanned using a gel doc (Supplementary Fig. 1).
Seeds of cv. 7042S were surface sterilized (0.02% mercuric chloride for 5 min), thoroughly rinsed in sterile distilled water and were soaked in LPS solution (SLPS) at the concentration of 50 µg/ml. In another set, to investigate the involvement of nitric oxide (NO) on LPS-induced defense response, SLPS procedure was followed by the treatment with NO scavenger 25 mM cPTIO (SLPS + cPTIO) 1 h prior to challenge inoculation. The suspensions were incubated at 27 °C in a rotary shaker for 6 h. Later, the seeds were allowed to dry in an incubator at 30 °C. Resistant (R) and Susceptible (S) seeds soaked in sterile distilled water for the same time duration served as positive control and negative control, respectively.
Pathogen inoculation and sampling of seedlings
The treated seeds were plated on moist blotters in Perspex plates and incubated for 2 days. The emerging seedlings were inoculated with the S. graminicola zoospore suspension of 4 × 104/ml concentration following the root-dip inoculation method and incubated at 25 ± 1 °C in dark. Another set of treated seedlings were not inoculated and were used as uninoculated controls. Seedlings were harvested at 0, 3, 6, 9, 12, 24, 48, and 72 h after inoculation (hai), wrapped immediately in aluminium foil, and stored at − 80 °C until further use for biochemical and molecular studies.
Estimation of nitric oxide
One gram of the harvested pearl millet seedlings was ground and homogenized in 1 ml buffer [0.1 M sodium acetate, 1 M NaCl, and 1% (w/v) ascorbic acid, pH 7.0]. The homogenate obtained was incubated immediately with 10 mM diaminofluorescein-FM (DAF-FM) for 1 h, followed by centrifugation at 10,000 rpm at 4 °C for 10 min. The resultant supernatant was collected and used for measurement of NO. Diaminofluorescein-2T (DAF-2T) is the reaction product of DAF with NO, which was measured by spectrofluorimeter with excitation and emission wavelengths of 495 and 515 nm, respectively (Kojima et al. 2001).
Histological studies
Time-course analysis of callose deposition
Callose deposition in pearl millet seedlings was analyzed according to the method described by Jensen (1962). The epidermal peelings were soaked in water-soluble aniline blue (0.005%) in 0.15 M dipotassium phosphate for 1 h. The stained peelings were mounted in glycerol and observed under fluorescence microscope, where k = 365–405 nm. Fluorescence was observed along the walls of the cells with callose deposition. Microscopic evaluation: in each case, 20 microscopic fields were counted for percentage calculation. The experiment was done in four replicates, each with 25 seedlings each and repeated 3 times. The peelings were examined under 500× and 1250× magnification for counting and photography, respectively.
Time-course analysis of the localization of H2O2
Hydrogen peroxide localization in pearl millet seedlings was analyzed. The epidermal peelings were soaked in 3,3′-diaminobenzidine (DAB, 1 mg/ml), pH 3.8. The stained peelings were mounted in 30% glycerol, 30% lactic acid, and observed under a microscope. Regions stained with the reddish-brown color indicated the accumulation of H2O2. In each case, 20 microscopic fields were recorded. The experiment was done in four replicates, with 25 seedlings each and repeated 3 times. The peelings were examined under 500× and 1250× magnification for counting and photography, respectively.
Biochemical studies
Enzyme assays
Enzyme extraction: harvested pearl millet seedlings (1 g fresh weight) were finely ground to a paste in 1 ml of extraction buffer. The extract was centrifuged at 12,000g for 20 min at 4 °C and the supernatant was transferred to a new tube and used as the enzyme extract.
Protein estimation
To calculate the specific activities of the enzymes, protein content in the crude extract was estimated by Lowry’s method (Lowry et al. 1951) using bovine serum albumin (Sigma) as a standard.
Phenylalanine ammonia-lyase (PAL) assay
One gram of the harvested pearl millet seedlings was ground to a fine paste in 25 mM Tris HCl buffer (pH 8.8). The activity of the PAL enzyme was assayed following the method described earlier (Beaudoin-Eagan and Thorpe 1985). 100 µl of enzyme extract was mixed with 900 µl of 50 mM L-phenylalanine and 100 mM Tris–HCl buffer solution and kept in a water bath at 40 °C for 120 min. The reaction was stopped by adding 60 µl of 5N HCl. PAL activity was determined as the amount of t-cinnamic acid formed from L-phenylalanine per mg of protein per min measured spectrophotometrically at a wavelength of 290 nm.
Peroxidase (POX) assay
One gram of the harvested pearl millet seedlings was ground to a fine paste in 1 ml of 10 mM potassium phosphate buffer (pH 6.9) and the extract was centrifuged at 12,000g for 20 min at 4 °C and the supernatant was transferred to a new tube and used as the enzyme extract. Peroxidase activity was assayed following the previously described method (Hammerschmidt and Nicholson 1999). The reaction mixture (3 ml) consisted of 0.25% v/v guaiacol in 10 mM potassium phosphate buffer (pH 6.0) containing 100 mM hydrogen peroxide. The crude enzyme extract (10 µl) was added to initiate the reaction, which was measured spectrophotometrically at 470 nm.
Hydroxyproline-rich glycoprotein (HRGP) assay
Preparation of cell wall and extraction of hydroxyproline (Hyp): extraction of pearl millet coleoptiles cell walls was carried out using the modified procedure of York et al. (1986). The roots and coleoptile regions of pearl millet seedlings were separated and homogenized using pestle and mortar at 4 °C in 0.5M potassium phosphate buffer (pH 7.0). Complete disruption of the cells was confirmed by observing the homogenate under a microscope. The broken cell suspension was centrifuged at 2000g for 10 min. Cell-wall preparation was washed repeatedly with the above buffer followed by sterile distilled water. Thereafter, cell walls were vigorously stirred and suspended in 5 volumes of 1:1 chloroform–methanol. Subsequently, the organic solvent was carefully removed. Cell walls were repeatedly washed with 5 volumes of acetone and then air-dried to obtain the cell-wall pellet. The Hyp content in the cell-wall hydrolysate was analyzed to determine the amount of HRGPs. The hydrolysis of the cell wall was carried out in the sealed tube using 6N HCl for 18 h at 110 °C. The hydrolysates were evaporated to dryness to remove traces of HCl. Hyp was then extracted in a minimum amount of sterile distilled water from the dried hydrolyzed samples and estimated Hyp as described previously (Prockop and Udenfriend 1960). Hyp content was expressed as µg Hyp/mg cell wall (dry weight).
Gene expression studies
Quantitative real‑time PCR analysis (qPCR)
The relative quantitation of PAL (NM_001174615.1), POX (EU492461), PR-1 (HQ699781.1), PR-5 (EU725133.1), and HRGP (GQ223398) mRNAs in harvested pearl millet seedling samples was done using gene-specific primers designed with Primer Express version 3.0 software (Applied Biosystems) with PP2A (protein phosphatase 2A) as endogenous reference gene (Siddaiah et al. 2017) (Table 1). The primer specifications were confirmed by running an agarose gel electrophoresis. Every qPCR reaction (20 µL) had 1 × SYBR Green PCR master mix (SYBR Green mix, Applied Biosystems), 3 pmol primers and 20 ng cDNA and used StepOnePlus™ Real-Time PCR Systems (Applied Biosystems). The qPCR steps included denaturation for 10 min at 95 °C, 40 cycles of 15 s at 95 °C, 60 s at 60 °C. A melting curve was created using a single cycle consisting of 15 s at 95 °C and 60 s at 60 °C at the end of each reaction. This was followed by a slow temperature increase to 95 °C at the rate of 0.3 °C/s. The quantification of target mRNAs used a comparative Ct method (Livak and Schmittgen 2001).
Table 1.
Primer sequences used for qRT-PCR amplification
| Sl. no. | Target gene | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) |
|---|---|---|---|
| 1 | PAL | ATGGAGTGCGAGAACGGCC | CTGCGCGATGCTGAGGCT |
| 2 | POX | CCCCAGAAGCACATTTGTGA | CATGGCTGCGGGCGGAG |
| 3 | PR-1 | TGGACGTGCCGCTGCCG | GAACTGCGCCGCCACACG |
| 4 | PR-5 | GCGTCCTCGGTCCTCCTG | CACACGCGGCCGGAGCTG |
| 5 | HRGP | GCCTAAGCCGAAGCCACCAA | GCGTGTAGGTCGGAGGAGTT |
| 6 | PP2A | TGAGAGCAGACAAATCACTCAA | AAGAGCTGTGAGAGGCAAATAA |
Effect of LPS pre-treatment on downy mildew disease incidence under greenhouse conditions
Pearl millet seeds treated with the inducers as described above were sown in earthen pots containing autoclaved soil, sand, and manure in the ratio 2:1:1 which were maintained at 25–30 °C with 95% relative humidity. The experiment consisted of four replicates for each treatment and the pots were arranged in a complete randomized block design. Seedlings were watered and fertilized when required. Seeds treated with sterile distilled water served as control and the seeds treated with systemic fungicide Metalaxyl formulation Apron 35SD at 6 g/kg served as a chemical control. The emerging 2-day-old seedlings were inoculated with downy mildew pathogen by whorl inoculation method (Singh and Gopinath 1985) with the suspension of S. graminicola zoospores at a concentration of 4 × 104/ml. The inoculated plants were observed for downy mildew disease expression and rated diseased when they expressed typical downy mildew symptoms like abaxial leaf sporulation, chlorosis, stunting, and earheads malformation. The downy mildew screening data were consolidated at 60 days after sowing (DAS).
Effect of LPS pre-treatment on downy mildew disease incidence under field conditions
The field experiment was conducted to determine the effect of LPS on pearl millet downy mildew disease incidence. Field trials were conducted in downy mildew nursery at the University of Mysore, during 2015–2016. The downy mildew nursery was a sick plot which was naturally infested with oospores of S. graminicola, which served as the source of primary inoculum. Infector rows raised 21 days prior to the raising of the test rows provided the additional inoculum (Williams 1984). Inducer treatments and controls were the same as described above. Treated seeds were hand-sown with a minimum of four replications per treatment. Each replication was a single row of 5 m length, hand seeded with 100–150 seeds per row. The read loamy soil of the field was watered adequately when required and the thinning of the plants was done after 21 days. The downy mildew disease was rated as described for greenhouse studies.
Statistical analysis
All experiments were carried out in four replicates. Data were analyzed separately for each experiment and were subjected to arcsine transformation and analysis of variance was carried out with transformation values (JMP Software; SAS Institute Inc., Cary, NC). The significance of the effect of treatments was determined by the magnitude of the F value (p ≤ 0.05). Tukey’s HSD test was applied for the separation of treatment means.
Results
LPS treatment enhances NO generation
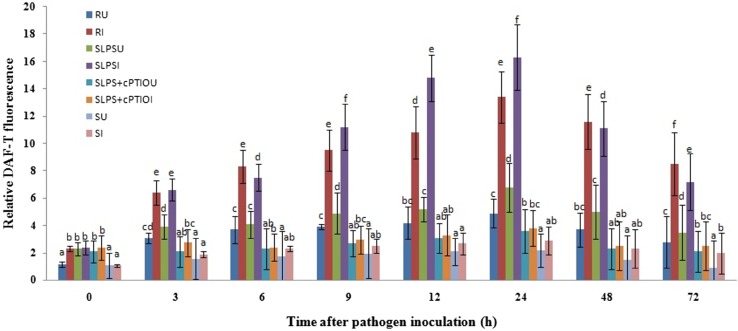
NO is suggested to play an important role during the induction of resistance in plants and it is also demonstrated that NO is involved in induced resistance in pearl millet against downy mildew. Therefore, to analyze the role of NO in LPS-induced resistance in pearl millet, a time course study was conducted. NO generation was low in all the categories of seedlings with or without pathogen inoculation. However, in all categories of inoculated seedlings, NO generation was significantly higher with time advancement in comparison with the uninoculated seedlings (Fig. 1). NO concentration was high in resistant seedlings compared to SLPS seedlings up to 6 h. NO levels were significantly higher in SLPS seedlings between 6 and 24 h compared to the resistant seedlings, beyond which again there was a fall in the levels of NO in SLPS seedlings. However, without inoculation, NO level in SLPS seedlings was considerably higher than that of resistant, SLPS + cPTIO treated and susceptible seedlings. In all categories of seedlings, NO concentration was peaked at 24 h with or without inoculation. The highest concentration of NO (16.3 nM) was recorded in SLPS seedlings at 24 hai which was 1.21, 4.27, and 5.62 folds higher than that of resistant, SLPS + cPTIO treated and susceptible seedlings, respectively. The concentration of NO was significantly lower in susceptible seedlings compared to the resistant and SLPS, SLPS + cPTIO treated seedlings with or without pathogen inoculation at all timepoints. These results demonstrated that NO generation is significantly enhanced due to LPS treatment in pearl millet.
Fig. 1.
Time course analysis of nitric oxide generation pattern in resistant (R), SLPS, SLPS + cPTIO treated and control/susceptible (S) pearl millet seedlings with (I) with or without (U) Sclerospora graminicola inoculation. All NO estimation experiments were carried out in four replicates. Vertical bars indicate standard error. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05
Histological studies
LPS treatment enhances callose deposition
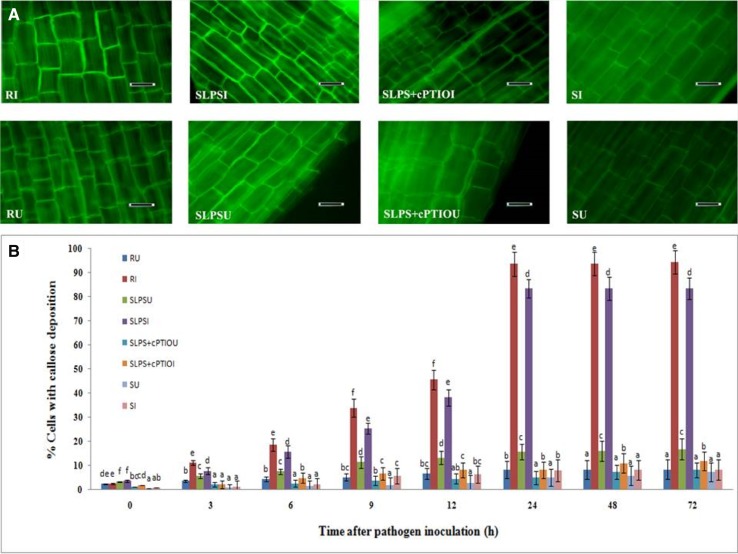
One of the early mechanisms of LPS elicited defense responses in plants is the alteration of the host cell wall at the site of infection to inhibit the entry of pathogen. Callose deposition is an important cell wall reinforcement process triggered by LPS and NO has a major role in callose deposition. Therefore, to elucidate the role of LPS in pearl millet cell-wall strengthening, callose deposition was studied in the treated and untreated seedlings. Callose deposition at basal level was observed in inoculated and uninoculated seedlings of all the groups. Among the uninoculated seedlings, maximum callose deposition was observed in SLPS seedlings compared to the uninoculated resistant, SLPS + cPTIO, and susceptible seedlings (Fig. 2a). After inoculation, callose deposition was gradually increased, and at 24 hai, a total of 93.7% and 83.5% cells with callose deposition was observed in resistant and SLPS-treated seedlings, respectively. At 24 hai, callose deposition in SLPS seedlings was 10.3 and 10.43 folds higher than that of SLPS + cPTIO and susceptible control seedlings respectively (Fig. 2b). Callose deposition results indicated that LPS treatment to pearl millet seeds increases the deposition of callose, particularly after downy mildew pathogen inoculation, in comparison with the untreated control.
Fig. 2.
a Light microscopic (fluorescence) pictures showing the deposition of callose in epidermal peelings from the coleoptile region pearl millet seedlings 24 h time interval with or without Sclerospora graminicola inoculation. Callose deposition detected by aniline blue staining method. b Temporal pattern of the degree of callose deposition in the coleoptile tissues of pearl millet seedlings at different time intervals with (I-inoculated) or without (U-uninoculated) Sclerospora graminicola inoculation. Results are average of three independent experiments with four replicates of 25 seedlings each. Vertical bars indicate standard error. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05. R—Seedlings of downy mildew resistant cultivar IP 18292, SLPS—downy mildew susceptible pearl millet seeds 7042S treated with LPS, SLPS + cPTIO—downy mildew susceptible pearl millet seeds 7042S treated with nitric oxide scavenger cPTIO prior to LPS treatment, S—downy mildew susceptible pearl millet seeds 7042S treated with SDW
LPS treatment increases H2O2 accumulation
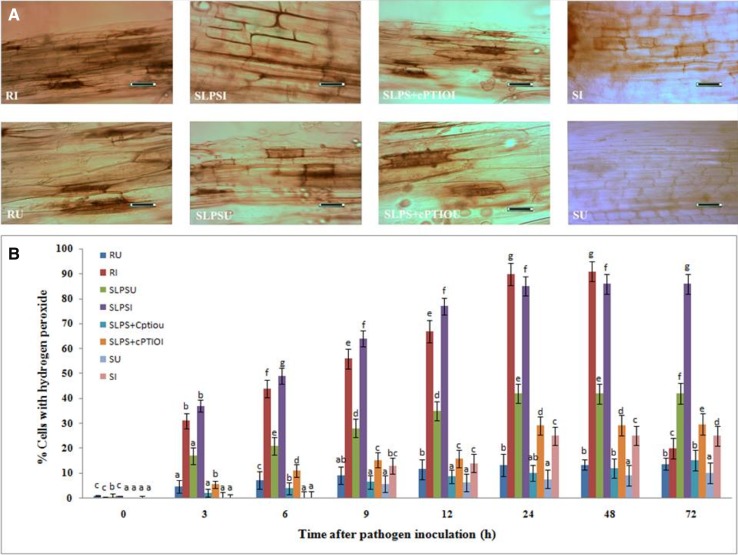
It has been demonstrated that H2O2 accumulation is vital for activation of various defense responses as it is a pivotal early signaling molecule, and therefore, to test whether H2O2 is involved in LPS elicited systemic resistance in pearl millet against downy mildew, we measured the H2O2 accumulation in test seedlings. The basal level of H2O2 accumulation was observed in all categories of seedlings with or without pathogen inoculation (Fig. 3a). Evidently, the concentration of H2O2 gradually increased up to 24 h after pathogen inoculation and plateaued thereafter. Maximum H2O2 accumulation was observed in resistant seedlings followed by SLPS seedlings. At 24 hai, H2O2 accumulation in SLPS seedlings was 2.91 and 3.4 folds higher than that of SLPS + cPTIO and susceptible seedlings, respectively (Fig. 3b). In SLPS + cPTIO treated seedlings, maximum accumulation was observed at 72 hai. The results showed that LPS treatment triggers the accumulation of H2O2 and scavenging of NO in LPS-treated seedlings significantly reduce the H2O2 accumulation.
Fig. 3.
a Light microscopic (bright field) pictures showing the accumulation of hydrogen peroxide in epidermal peelings from the coleoptile region pearl millet seedlings 24 h time interval with or without Sclerospora graminicola inoculation accumulation of hydrogen peroxide was detected by DAB staining method. b Temporal pattern of the degree of accumulation of hydrogen peroxide in the coleoptile tissues of pearl millet seedlings at different time intervals with (I-inoculated) or without (U-uninoculated) Sclerospora graminicola inoculation. Results are average of three independent experiments with four replicates of 25 seedlings each. Vertical bars indicate standard error. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05. R—Seedlings of downy mildew resistant cultivar IP 18292, SLPS- downy mildew susceptible pearl millet seeds 7042S treated with LPS, SLPS + cPTIO − downy mildew susceptible pearl millet seeds 7042S treated with nitric oxide scavenger cPTIO prior to LPS treatment, S—downy mildew susceptible pearl millet seeds 7042S treated with SDW
Biochemical studies
LPS treatment triggers increased activities of defense enzymes and HRGPs
PAL activity
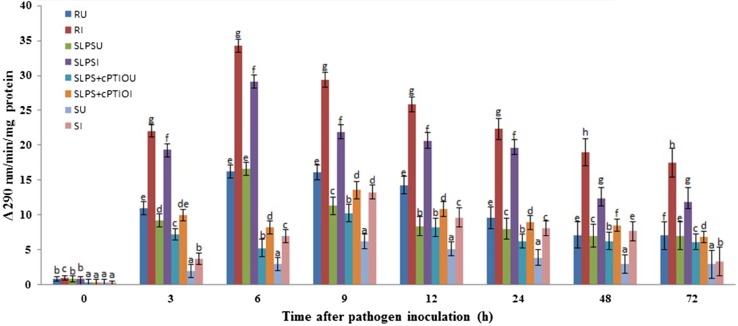
Constitutive PAL activity was observed in resistant, SLPS and susceptible seedlings with or without pathogen inoculation. The activity was significantly increased in a time-dependent manner. The constitutive PAL activity was significantly lesser in SLPS + cPTIO and susceptible seedlings compared to resistant and SLPS seedlings with or without pathogen inoculation. At all the tested timepoints, PAL activity was higher in resistant seedlings compared to SLPS, SLPS + cPTIO, and susceptible seedlings with or without pathogen inoculation. In both inoculated and uninoculated samples, the activity was peaked at 6 hai in resistant and SLPS seedlings, whereas in SLPS + cPTIO and susceptible seedlings, maximum activity was observed at 9 hai. In resistant seedlings, the highest activity of 34.2 units was recorded at 6 hai. In SLPS-treated seedlings, maximum PAL activity of 29.1 units was recorded at 6 hai which was 3.54 and 4.20 folds higher than that of SLPS + cPTIO and susceptible at the corresponding timepoint. Among the uninoculated seedlings, PAL activity in SLPS was on par with resistant seedlings at 6 hai and significantly higher than that of SLPS + cPTIO and susceptible seedlings. The treatment with cPTIO before inoculation with pathogen resulted in decreased PAL activity at 3 hai. At all the timepoints, enzyme activity was significantly lower in susceptible seedlings compared to the resistant and LPS-treated seedlings (Fig. 4). Overall, PAL activity is enhanced significantly in LPS-treated seedlings, particularly after pathogen inoculation, and NO was demonstrated to be responsible for the increased PAL activity and scavenging of NO in the treated seedlings resulted in the reduced enzyme activity.
Fig. 4.
Time course analysis of PAL activity in resistant (R), LPS treated (SLPS), LPS treatment plus NO scavenger treatment (SLPS + cPTIO) and control/susceptible (S) pearl millet seedlings with (I) or without (U) S. graminicola Inoculation. PAL activity was determined as the amount of t-cinnamic acid formed from l-phenylalanine per mg of protein per min measured spectrophotometrically at a wavelength of 290 nm. Results are average of three independent experiments each carried out in four replicates. Vertical bars indicate standard error. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05
POX activity
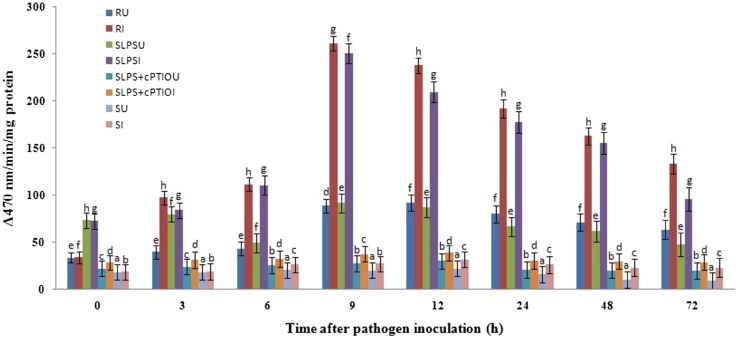
POX is known to have a multifaceted role in plant defense including lignification, cross-linking of phenolics and glycoproteins, suberization, and phytoalexin production. To determine the role of POX in LPS-induced resistance in pearl millet, POX assay was carried out in the test seedlings. Constitutive peroxidase activity was recorded in resistant, LPS treated and susceptible seedlings which gradually and significantly increased in a time-dependent manner. At all the tested timepoints, POX activity was higher in resistant seedlings compared to SLPS and susceptible seedlings. In resistant and SLPS treated seedlings, maximum POX activity was observed at 9 hai, whereas in SLPS + cPTIO and susceptible seedlings, the enzyme activity peaked at 12 hai with or without pathogen inoculation. In resistant seedlings, the highest activity of 74.68 units was recorded at 9 h after inoculation. In SLPS-treated seedlings, maximum POX activity of 71.63 unit was recorded at 9 hai which was 6.72 and 9.68 folds higher than that of SLPS + cPTIO and susceptible seedlings, respectively. The treatment with cPTIO before inoculation with pathogen resulted in decreased POX activity after 3 hai. At all the timepoints, the enzyme activity was significantly lower in susceptible seedlings compared to the resistant and LPS-treated seedlings (Fig. 5). These results indicated that LPS treatment induces POX activity through the generation of NO.
Fig. 5.
Time course analysis of POX activity in resistant (R), LPS treated (SLPS), LPS treatment plus NO scavenger treatment (SLPS + cPTIO) and control/susceptible (S) pearl millet seedlings with (I) or without (U) S. graminicola Inoculation. Peroxidase activity determined as the increase in absorbance recorded 470 nm. POX activity is expressed in terms of the change in A470/min/mg protein. Results are average of three independent experiments each carried out in four replicates. Bars indicate ± SE. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05
HRGPs analysis
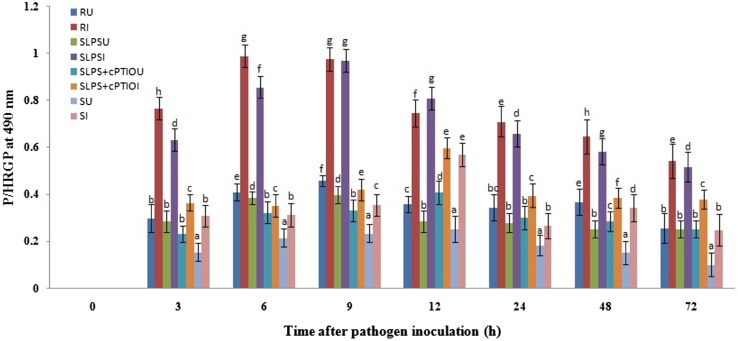
Increased cross-linking of cell-wall proteins particularly hydroxyproline-rich glycoproteins is known to be one of the important cell-wall reinforcement mechanisms of resistance inducing elicitors or inducers. Therefore, to understand the role of HRGPs in LPS-induced resistance in pearl millet, levels of HRGPs were analyzed in the test seedlings. Constitutive HRGPs activity was observed in resistant, SLPS, and susceptible seedlings which gradually and significantly increased with time. At all the tested timepoints, HRGPs level was higher in resistant seedlings compared to SLPS, SLPS + cPTIO, and susceptible seedlings. The maximum concentration of HRGPs was observed at 9 hai in resistant and SLPS treated seedlings, whereas SLPS + cPTIO and susceptible seedlings displayed maximum concentration at 12 hai in. At 9 hai, the concentration of HRGPs was 0.976 µg Hyp/mg cell wall (dry weight) in resistant seedlings which were on par with SLPS treated seedlings (0.971 µg Hyp/mg cell wall [dry weight]). HRGPs concentration in SLPS-treated seedlings at 9 hai was 2.31 and 2.73 folds higher compared to SLPS + cPTIO and susceptible seedlings. Treatment with cPTIO before inoculation with pathogen resulted in a decreased concentration of HRGPs at 3 hai. At all the timepoints, HRGPs were significantly lower in susceptible seedlings compared to the resistant and LPS treated seedlings (Fig. 6). A significant increase in HRGPs concentration in LPS-treated seeds in comparison with the untreated control and these results indicated that cell-wall strengthening by crosslinking of HRGPs is induced by LPS treatment.
Fig. 6.
Temporal pattern of accumulation of HSPGs in 2-day-old pearl millet seedlings harvested at 0, 3, 6, 9, 12, 24, 48, and 72 h with (I-inoculated) or without (U-uninoculated) Sclerospora graminicola inoculation. R—Seedlings of downy mildew resistant cultivar 18292, SLPS—downy mildew susceptible pearl millet seeds 7042S treated with LPS, SLPS + cPTIO—downy mildew susceptible pearl millet seeds 7042S treated with nitric oxide scavenger cPTIO prior to LPS treatment, S—downy mildew susceptible pearl millet seeds 7042S treated with SDW. Hydroxyproline (Hyp) content was expressed as µg Hyp/mg cell wall (dry weight). Results are average of three independent experiments each carried out in four replicates. Vertical Bars indicate standard error. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05
Gene expression studies
LPS treatment upregulates defense genes expression
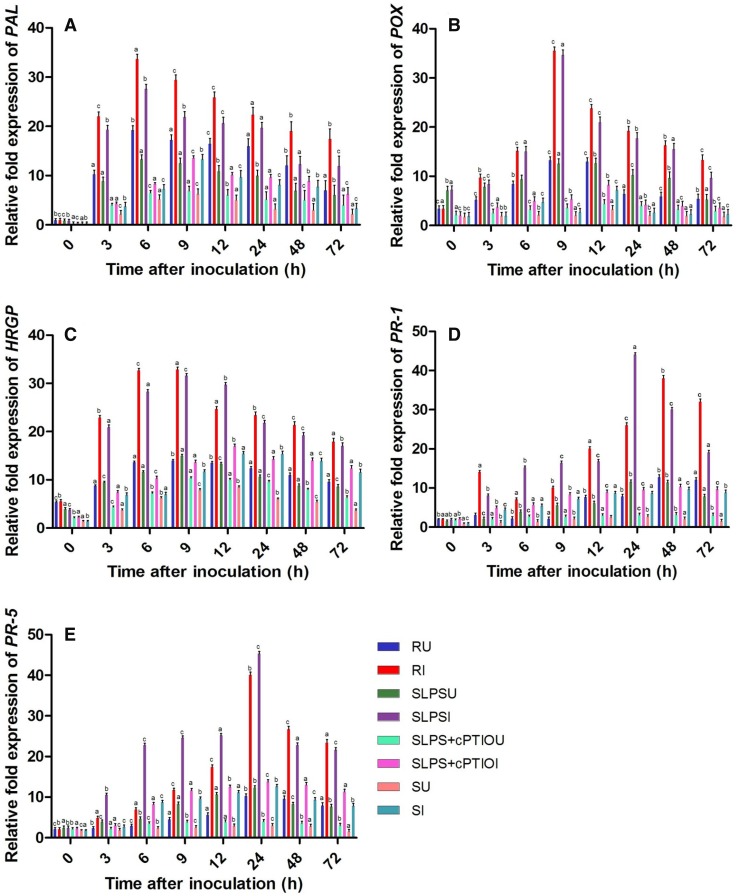
Induction of resistance is dependent on a coordinated expression of a set of genes which majorly encodes defense enzymes and proteins, and pathogenesis-related (PR) proteins. To study the gene expression of major defense-related proteins (PR-1, PR-5, HRGPs) and enzymes (PAL, POX) in the test seedlings, real-time PCR analysis was performed. Basal levels of PAL transcripts were detected in all categories of seedlings with or without pathogen inoculation and the expression levels were higher in resistant and SLPS seedlings compared to the susceptible controls. In all sets of seedlings, PAL gene expression was higher in inoculated samples compared to the uninoculated samples at all the tested timepoints. The expression of PAL gene was highest at 6 hai in resistant and SLPS seedlings, whereas in susceptible seedlings, maximum PAL gene expression was recorded at 9 hai. The PAL gene expression was highest in the resistant seedlings at all timepoints compared to the SLPS and susceptible seedlings with or without pathogen inoculation. In SLPS-treated seedlings, PAL expression at 6 hai was 3.33 and 3.82 folds higher than that of SLPS + cPTIO treated and susceptible seedlings at the same timepoint. In uninoculated SLPS-treated seedlings, PAL expression at 6 h was 2.03 and 2.58 folds higher than the SLPS + cPTIO treated and control seedlings (Fig. 7a).
Fig. 7.
qRT-PCR determined the relative expression of genes of various defense enzymes and proteins in 2-day-old pearl millet seedlings with (I) or without (U) Sclerospora graminicola inoculation harvested 0, 3, 6, 9, 12, 24, 48, and 72 h. R—Seedlings of downy mildew resistant cultivar IP 18292, SLPS—downy mildew susceptible pearl millet seeds 7042S treated with LPS, SLPS + cPTIO—downy mildew susceptible pearl millet seeds 7042S treated with nitric oxide scavenger cPTIO prior to LPS treatment, S—downy mildew susceptible pearl millet seeds 7042S treated with sterile distilled water. a Phenylalanine ammonia lyase genes b POX genes, c HRGPs genes d PR-1 genes, and e PR-5 genes. Values are means of a single experiment carried out in triplicate. The bars indicate ± SE and the data were analyzed by one-way ANOVA followed by Tukey’s test and p value < or = 0.05 was significant compared with control and < 0.01 significant with treated control
The increased amount of peroxidase transcripts was detected at all time intervals in resistant, SLPS treated and susceptible seedlings with or without inoculation. However, POX gene expression was higher in inoculated samples compared to the uninoculated counterpart. In resistant and SLPS treated seedlings, POX expression was peaked at 9 h with or without inoculation as against the susceptible seedlings where POX expression was peaked at 12 h with or without inoculation. Maximum POX expression was recorded at 9 hai, and at this timepoint, POX gene expression in SLPS-treated seedlings was on par with resistant seedlings, and 6.49 and 13.11 folds higher than the SLPS + cPTIO treated and susceptible control seedlings, respectively (Fig. 7b).
Constitutive expression of HRGP transcripts was evident in all categories of seedlings with or without inoculation and highest was observed in resistant seedlings. However, there was a marked increase in the HRGP transcript with time advancement, and the expression peaked at 9 h in resistant and SLPS seedlings, whereas, in susceptible seedlings, expression peaked at 12 h. At 9 h timepoint, HRGPs expression in inoculated SLPS seedlings was 2.32, and 2.72 folds higher than that of SLPS + cPTIO treated and susceptible controls, respectively. At 12 hai, HRGPs’ expression in SLPS-treated seedlings was 1.21 folds higher than the resistant seedlings (Fig. 7c).
PR proteins are an integral part of the plant defense system and their expression is upregulated during pathogen infection to elicit defense responses. Hence, we have dissected the expression of PR transcripts particularly PR-1 and PR-5 in the test seedlings. The increased amount of PR-1 transcript was noticed in all categories of seedling at the constitutive level, which was increased in a time-dependent manner. In all the categories of seedlings, PR-1 gene expression was higher in inoculated samples compared to the uninoculated counterpart at all timepoints. It was noted that PR-1 expression peaked at 24 h time point in SLPS-treated seedlings, whereas in resistant, SLPS + cPTIO treated and susceptible controls maximum expression was at 48 h, with or without inoculation. PR-1 expression in SLPS-treated seedlings was 1.69, 4.63, and 5.12 folds higher than resistant, SLPS + cPTIO treated, and control seedlings at 24 h, respectively. PR-1 expression in inoculated SLPS seedlings was 3.82, 5.11, and 16.29 folds higher than that of uninoculated SLPS, inoculated susceptible, and uninoculated susceptible seedlings at 24 h timepoint, respectively (Fig. 7d).
Similarly, PR-5 transcript accumulation was noticed in all categories of seedling at the constitutive level, which increased with time. In all seedlings, with or without pathogen inoculation, PR-5 gene expression peaked at 24 h timepoint. PR-5 expression was higher in SLPS-treated seedlings compared to both resistant and control seedlings between 3 and 24 h. Maximum PR-5 expression was observed at 24 hai in SLPS-treated seedlings which were 1.13 folds higher than resistant seedlings. PR-5 expression in inoculated SLPS seedlings was 3.28 and 3.59 folds higher than that of SLPS + cPTIO treated and susceptible seedlings at 24 h timepoint, respectively. However, PR-5 expression was higher in resistant seedlings compare to SLPS treated and control seedlings from 48 h onwards (Fig. 7e). qPCR results demonstrated that LPS treatment is responsible for the increased expression of plant defense-related genes. The regulation of these genes is modulated by the enhanced generation of NO.
LPS induces resistance against downy mildew disease
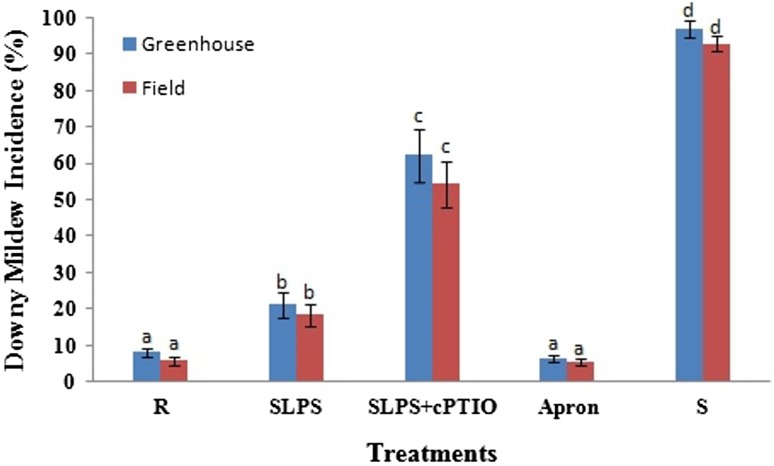
A series of greenhouse and field experiments were conducted to demonstrate the ability of LPS to protect pearl millet plants against downy mildew disease and also to examine the role of NO in LPS mediated induced resistance. Downy mildew disease incidence was significantly reduced due to treatment both under greenhouse and field conditions. Furthermore, it was noted that SLPS + cPTIO treatment drastically increased downy mildew disease incidence suggesting the possible role for NO in LPS-mediated resistance. Among all the treatments, Apron showed the lowest downy mildew incidence both under greenhouse and field conditions (Fig. 8).
Fig. 8.
Illustration of downy mildew disease incidence in resistant (R), LPS treated (SLPS), LPS treatment plus NO scavenger treatment (SLPS + cPTIO) and control/susceptible (S) pearl millet seedlings under (a) greenhouse and (b) field conditions
Under greenhouse conditions, a significant reduction of downy mildew disease was observed in resistant seedlings which showed the disease incidence of 8.2%. SLPS and SLPS + cPTIO treatments recorded downy mildew incidence of 21.3 and 62.3%, respectively, as against susceptible control which recorded 97% downy mildew incidence (Fig. 9).
Fig. 9.
Downy mildew disease incidence in resistant (R), LPS treated (SLPS), LPS treatment plus NO scavenger treatment (SLPS + cPTIO) and control/susceptible (S) pearl millet seedlings with S. graminicola inoculation. Greenhouse and field experiments were carried out in four replicates. Vertical bars indicate standard error. Means designated with the same letter are not significantly different according to Tukey’s HSD at p ≤ 0.05
Under field conditions, maximum downy mildew reduction was in resistant seedlings which showed 5.9% downy mildew incidence. SLPS and SLPS + cPTIO treatments recorded 18.6 and 54.4% downy mildew as against susceptible control which showed 93% downy mildew incidence (Fig. 9). The downy mildew reaction studies, both under greenhouse and field conditions, clearly showed that LPS treatment is effective in managing downy mildew disease by inducing systemic resistance in the host and such elicitation of resistance is likely to be mediated by the enhanced generation of NO.
Discussion
Elicitor signaling primes the host defense responses in plants and effectively offers resistance against pathogens by mechanisms such as cell-wall reinforcement, production of antimicrobial metabolites, expression of defense enzymes and proteins, and hypersensitive responses (Lorrain et al. 2003; Lavanya et al. 2017). LPS is an elicitor of systemic resistance in many crops. LPS has been known to possess diverse effects in plants including synthesis of NO, phosphorylation of MAPKs, and priming for more effective induction of various defense responses (Felix et al. 1999; Dow et al. 2000; Gerber et al. 2004; Desaki et al. 2006; Erbs and Newman 2012; Newman et al. 2007). In this study, we examined the possible mechanisms underlying LPS induced resistance in pearl millet against the oomycete Sclerospora graminicola by analyzing the role of various defense enzymes and PR proteins. Furthermore, we attempted to elucidate the role of NO in LPS mediated resistance induction.
Nitric oxide is a vital signaling compound which acts as an early messenger which in turn primes other signaling events (Garcia-Brugger et al. 2006; Siddaiah et al. 2017). The time course analysis of LPS-treated pearl millet seedlings upon inoculation with Sclerospora graminicola, recorded earlier generation and significant elevation in NO levels as compared to the untreated seedlings. It has been previously shown that NO production plays a vital role during innate-immune responses triggered by LPS (Melotto et al. 2006). In Arabidopsis thaliana, LPS preparations from different bacteria triggered innate immunity mediated by NO, indicating that NO is an early mediator during LPS treatment (Zeidler et al. 2004). In Arabidopsis thaliana, it was shown that enhanced NO production is one of the immediate responses following LPS perception leading to defense gene induction (Sun et al. 2012). Endogenous NO production was recorded in plant cells challenged by avirulent pathogens and elicitors (Wendehenne et al. 2004; Delledonne 2005) emphasizing that NO is a part of intracellular signaling cascades activated in plant cells in response to pathogens or elicitors.
The histological studies in our experiments showed that LPS-induced resistance is mediated by NO and is associated with cell wall modifications like callose deposition and H2O2 accumulation. Both callose deposition and H2O2 accumulation occurred quicker and in higher rate in SLPS seedlings compared to the other checks with or without pathogen inoculation. Furthermore, in the presence of NO scavenger cPTIO, these cell-wall modifications were slowed and weakened confirming the role of NO in these defense responses. Our results corroborate earlier reports stating that cell-wall strengthening is an important aspect of induced resistance, and NO plays an important in elicitor-induced wall modifications. LPS elicited immunity modulates through NO production, induction of PR gene expression, and cell-wall alterations like deposition of callose and phenolics (Erbs and Newman 2012). Sun and Li (2013) demonstrated that, in Arabidopsis thaliana, LPS treatments induce an array of defense responses which are modulated by NO generation such as enhanced callose deposition (Sun and Li 2013). Pseudomonas fluorescens induced resistance against Rhizoctonia solani in bean was found to be mediated by NO which increased H2O2 production; while the use of NO scavenger cPTIO resulted in decreased resistance and H2O2 production (Keshavarz-Tohid et al. 2016). Increased cross-linking of cell-wall proteins and callose deposition during NO-mediated induced resistance in tomato against Colletotrichum coccodes and Rhizoctonia solani were reported (Wang and Higgins 2005; Noorbakhsh and Taheri 2016).
Furthermore, the present study also showed that NO production was a very vital response of LPS treatment to pearl millet and its production and concentration modulated various other defense responses including the production of defense enzymes, PR proteins, and HRGPs which ultimately led to elevated resistance against downy mildew disease. Furthermore, scavenging of NO in LPS-treated seedlings resulted in a significant decrease in all these defense responses implicating that NO is central to LPS elicited resistance.
In plants, PAL plays a key role in the synthesis of lignins and isoflavonoid phytoalexins, both of which are involved in plant defense reactions (Hahlbrock and Scheel 1989). In the present investigations, PAL enzyme activity and also gene expression were significantly enhanced in SLPS seedlings compared to untreated seedlings. Similar observations were reported in Arabidopsis thaliana treated with LPS derived from the bacterial pathogens Pectobacterium atrosepticum and Pectobacterium carotovorum subsp. carotovorum significantly induced earlier and higher expression of PAL transcripts (Mohamed et al. 2015). Furthermore, involvement of PAL during pearl millet—S. graminicola interaction has been well documented conferring a major role of this enzyme in resistance development (Nagarathna et al. 1993; Geetha et al. 2005). Furthermore, NO-induced PAL enzyme in pearl millet seedlings during the induced resistance against downy mildew disease (Manjunatha et al. 2009a). Likewise, there are several earlier reports which have confirmed the role of NO during enhanced activities of PAL in various host-pathogen systems (Bowler et al. 1994). NO-mediated plant defense activation significantly enhanced the expression of PAL in tobacco and wheat (Durner et al. 1998; de Pinto et al. 2002; Guo et al. 2004).
Defense enzymes, especially POX interferes with disease development and progression through the formation of polymerized phenolic barriers around the sites of infection (Smit and Dubery 1997; Li and Steffens 2002) and trigger the synthesis of anti-nutritive, antibiotic, and cytotoxic compounds leading to enhanced resistance against pathogens (Hammerschmidt and Nicholson 1999). In the current analysis, POX expression and its enzyme activity were investigated in resistant, SLPS and susceptible seedlings after pathogen inoculation at all the time points. However, the expression was higher in resistant and SLPS seedlings compared to susceptible seedlings, further confirming the significance of this enzyme in pearl millet downy mildew interaction. The role of POX as a marker of systemic acquired resistance has been well established in numerous resistance induction studies. However, its role in LPS induced resistance has been less reported. Nonetheless, the role of POX as an important defense enzyme in imparting host resistance by different inducers/elicitors against pearl millet downy mildew system is also well demonstrated (Pushpalatha et al. 2007; Deepak et al. 2007; Manjunatha et al. 2008; Raj et al. 2012). In addition, peroxidase seemed to be a vital enzyme whose levels were significantly enhanced during NO-mediated resistance induction in pearl millet against downy mildew disease (Manjunatha et al. 2009b). NO-mediated induced resistance against Rhizoctonia solani in bean correlated with enhanced peroxidase activities (Keshavarz-Tohid et al. 2016).
HRGPs are plant cell-wall structural components which are known to play a vital role in host defense responses towards pathogen invasion (Davies et al. 1997). Pathogen infections or pathogen-derived elicitor treatments have increased the level of HRGPs and subsequently induced resistance against various pathogens (Bradley et al. 1992; Brownleader et al. 1995; Kang and Buchenauer 2003). Different elicitors induced downy mildew resistance in pearl millet which correlated with increased HRGPs content in the cell wall; particularly, maximum HRGPs accumulation was observed during Pseudomonas fluorescens UOM SAR 14 treatment (Sujeeth et al. 2010; Siddaiah et al. 2018). It is interesting to note here that the LPS used in the present study was obtained from Pseudomonas fluorescens UOM SAR 14 which implied that LPS has a role in HRGPs accumulation. The role of NO as a key signal component in accumulation of HRGPs was also demonstrated by our earlier studies wherein priming of pearl millet seedlings with NO donors effectively induced hypersensitive reactions and enhanced accumulation of proline/hydroxyproline-rich glycoprotein during infection by downy mildew pathogen Sclerospora graminicola; and endogenous NO concentration regulated the degree of defense responses like hypersensitive reaction development, H2O2 accumulation, and HRGPs cross-linking (Manjunatha et al. 2009a).
Pathogenesis-Related (PR) proteins are a group of diverse proteins whose accumulation is triggered by pathogen attack or abiotic stress. PR-1 and PR-5 proteins are widely studied and are well established as markers of systemic acquired resistance in several host–pathogen systems. Moreover, PR-1 and PR-5 have been found to possess antifungal activity against oomycetes. In earlier studies involving induced resistance against pearl millet downy mildew disease, inducers like L-methionine, and Bacillus pumilus INR7 have shown accumulation of PR-1 and PR-5 corresponding with the increased development of resistance (Sarosh et al. 2005). Though there are several reports confirming the involvement of PR proteins during NO-mediated resistance in plants, the role of LPS during such resistance development is less reported. Our results corroborate earlier studies which have shown that LPS treatment primes the induction and expression of various PR proteins in different host–pathogen systems (Dow et al. 2000). Microarray studies in Arabidopsis thaliana plants showed that LPS treatment induced the expression of an array of defense- or stress-associated genes, including glutathione S-transferases, cytochrome P450, and many gene-encoding PR proteins, both locally and systemically (Zeidler et al. 2004). Furthermore, Zeidler et al. showed that NOS mutant Arabidopsis plants, even when treated with LPS, completely failed to express any defense-related genes, thus emphasizing that perception of LPS and induction of NOS contribute towards the activation of plant defense responses. Prior treatment of lipooligosaccharides from plant–pathogen Xanthomonas campestris pv. campestris induced the defense-related genes PR-1 and PR-2 in Arabidopsis (Silipo et al. 2005). In Arabidopsis, LPS treatment resulted in enhanced PR-1 gene expression and it was modulated by NO generation (Sun et al. 2012; Sun and Li 2013).
Conclusions
To the best our knowledge, this is the first report on the evaluation of expression and activity of various defense enzymes/proteins in the LPS-mediated induction of resistance in a monocot–oomycete system. For most of the enzyme activities and genes examined, the highest levels of activities and transcripts were observed in resistant seedlings followed by SLPS seedlings in comparison with the susceptible seedlings. Measurement of mRNA levels demonstrates that genes encoding POX, PAL, HRGPs, PR-1, and PR-5 were induced prominently by LPS treatment. The enzyme products of the genes examined are predicted to be involved in the biosynthesis of defense compounds, so it is not surprising that their transcripts increased following pathogen inoculation.
Enhanced accumulation of defense enzymes and PR proteins showed NO as the main gene induction signal. This result is indicative of NO as the main signal molecule triggered by LPS treatment, particularly during pearl millet–downy mildew host–pathogen interaction. Induced systemic resistance mediated by LPS has been demonstrated only in a few plant–pathogen systems and the biochemical and molecular mechanisms underlying this phenomenon are not investigated completely. Overall, the present study demonstrates the plausible involvement of the important defense enzymes like PAL, POX, PR proteins, and HRGPs leading to hypersensitive response during LPS-mediated induced resistance against pearl millet downy mildew and that NO is a central signal to all these defense manifestations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
LSN, UAC, NRS, and CNS conceived the project. CDM, GVK, TC, SR, and CNS designed the experiments. LSN, UAC, NRS, and CNS carried out the research and analysis of data. LSN, UAC, CDM, and CNS wrote the paper.
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Beaudoin-Eagan LD, Thorpe TA. Tyrosine and phenylalanine ammonia lyase activities during shoot initiation in tobacco callus cultures. Plant Physiol. 1985;78(3):438–441. doi: 10.1104/pp.78.3.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Van Camp W, Van Montagu M, Inze D, Asada K. Superoxide dismutase in plants. Crit Rev Plant Sci. 1994;13(3):199–218. doi: 10.1080/07352689409701914. [DOI] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70(1):21–30. doi: 10.1016/0092-8674(92)90530-P. [DOI] [PubMed] [Google Scholar]
- Brownleader MD, Ahmed N, Trevan M, Chaplin MF, Dey PM. Purification and partial characterization of tomato extensin peroxidase. Plant Physiol. 1995;109(3):1115–1123. doi: 10.1104/pp.109.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coventry HS, Dubery IA. Lipopolysaccharides from Burkholderia cepacia contribute to an enhanced defensive capacity and the induction of pathogenesis-related proteins in Nicotianae tabacum. Physiol Mol Plant Pathol. 2001;58(4):149–158. doi: 10.1006/pmpp.2001.0323. [DOI] [Google Scholar]
- Davies HA, Daniels MJ, Dow JM. Induction of extracellular matrix glycoproteins in Brassica petioles by wounding and in response to Xanthomonas campestris. Mol Plant Microbe Interact. 1997;10(7):812–820. doi: 10.1094/MPMI.1997.10.7.812. [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Tommasi F, De Gara L. Changes in the antioxidant systems as part of the signaling pathway responsible for the programmed cell death activated by nitric oxide and reactive oxygen species in tobacco Bright-Yellow 2 cells. Plant Physiol. 2002;130(2):698–708. doi: 10.1104/pp.005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepak S, Shailasree S, Kini RK, Hause B, Shetty SH, Mithöfer A. Role of hydroxyproline-rich glycoproteins in resistance of pearl millet against downy mildew pathogen Sclerospora graminicola. Planta. 2007;226(2):323–333. doi: 10.1007/s00425-007-0484-4. [DOI] [PubMed] [Google Scholar]
- Delledonne M. NO news is good news for plants. Curr Opin Plant Biol. 2005;8(4):390–396. doi: 10.1016/j.pbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Desaki Y, Miya A, Venkatesh B, Tsuyumu S, Yamane H, Kaku H, Minami E, Shibuya N. Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 2006;47(11):1530–1540. doi: 10.1093/pcp/pcl019. [DOI] [PubMed] [Google Scholar]
- Dow M, Newman M-A, von Roepenack E. The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Ann Rev Phytopathol. 2000;38(1):241–261. doi: 10.1146/annurev.phyto.38.1.241. [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci. 1998;95(17):1032810333. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs G, Newman M-A. The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Mol Plant Pathol. 2012;13(1):95–104. doi: 10.1111/j.1364-3703.2011.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18(3):265–276. doi: 10.1046/j.1365-313X.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B, Wendehenne D, Pugin A. Early signaling events induced by elicitors of plant defenses. Mol Plant Microb Interact. 2006;19(7):711–724. doi: 10.1094/MPMI-19-0711. [DOI] [PubMed] [Google Scholar]
- Geetha N, Amruthesh K, Sharathchandra R, Shetty HS. Resistance to downy mildew in pearl millet is associated with increased phenylalanine ammonia lyase activity. Funct Plant Biol. 2005;32(3):267–275. doi: 10.1071/FP04068. [DOI] [PubMed] [Google Scholar]
- Gerber IB, Zeidler D, Durner J, Dubery IA. Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia. Planta. 2004;218(4):647–657. doi: 10.1007/s00425-003-1142-0. [DOI] [PubMed] [Google Scholar]
- Guo J-H, Qi H-Y, Guo Y-H, Ge H-L, Gong L-Y, Zhang L-X, Sun P-H. Biocontrol of tomato wilt by plant growth-promoting rhizobacteria. Biol Control. 2004;29(1):66–72. doi: 10.1016/S1049-9644(03)00124-5. [DOI] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Ann Rev Plant Biol. 1989;40(1):347–369. doi: 10.1146/annurev.pp.40.060189.002023. [DOI] [Google Scholar]
- Hammerschmidt R, Nicholson RL. A survey of plant defense responses to pathogens. Induced plant defenses against pathogens and herbivores. St. Minn: APS Press; 1999. pp. 55–72. [Google Scholar]
- Jetiyanon K, Plianbangchang P. Lipopolysaccharide of Enterobacter asburiae strain RS83: a bacterial determinant for induction of early defensive enzymes in Lactuca sativa against soft rot disease. Biol Control. 2013;67(3):301–307. doi: 10.1016/j.biocontrol.2013.09.014. [DOI] [Google Scholar]
- Kang Z, Buchenauer H. Immunocytochemical localization of cell wall-bound thionins and hydroxyproline-rich glycoproteins in Fusarium culmorum-infected wheat spikes. J Phytopathol. 2003;151(3):120–129. doi: 10.1046/j.1439-0434.2003.00693.x. [DOI] [Google Scholar]
- Keshavarz-Tohid V, Taheri P, Taghavi SM, Tarighi S. The role of nitric oxide in basal and induced resistance in relation with hydrogen peroxide and antioxidant enzymes. J Plant Physiol. 2016;199:29–38. doi: 10.1016/j.jplph.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Lavanya SN, Niranjan-Raj S, Nayaka SC, Amruthesh KN (2017) Systemic protection against pearl millet downy mildew disease induced by cell wall glucan elicitors from Trichoderma hamatum UOM 13. J Plant Protect Res
- Li L, Steffens JC. Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta. 2002;215(2):239–247. doi: 10.1007/s00425-002-0750-4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 – ∆∆CT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balagué C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8(6):263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- Lyon G (2007) Agents that can elicit induced resistance. Induced resistance for plant defence: a sustainable approach to crop protection:9–29
- Manjunatha G, Raj SN, Shetty NP, Shetty HS. Nitric oxide donor seed priming enhances defense responses and induces resistance against pearl millet downy mildew disease. Pesticide Biochem Physiol. 2008;91(1):1–11. doi: 10.1016/j.pestbp.2007.11.012. [DOI] [Google Scholar]
- Manjunatha G, Deepak S, Geetha PN, Niranjan-Raj S, Kini RK, Shetty HS. Hypersensitive reaction and P/HRGP accumulation is modulated by nitric oxide through hydrogen peroxide in pearl millet during Sclerospora graminicola infection. Physiol Mol Plant Pathol. 2009;74(2):191–198. doi: 10.1016/j.pmpp.2009.12.001. [DOI] [Google Scholar]
- Manjunatha G, Niranjan-Raj S, Prashanth GN, Deepak S, Amruthesh KN, Shetty HS. Nitric oxide is involved in chitosan-induced systemic resistance in pearl millet against downy mildew disease. Pest Manag Sci. 2009;65(7):737–743. doi: 10.1002/ps.1710. [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126(5):969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mohamed K-H, Daniel T, Aurélien D, El-Maarouf-Bouteau H, Rafik E, Arbelet-Bonnin D, Biligui B, Florence V, Mustapha EM, François B. Deciphering the dual effect of lipopolysaccharides from plant pathogenic Pectobacterium. Plant Signal Behav. 2015;10(3):e1000160. doi: 10.1080/15592324.2014.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarathna K, SHETTY SA, Shetty HS. Phenylalanine ammonia lyase activity in pearl millet seedlings and its relation to downy mildew disease resistance. J Exp Bot. 1993;44(8):1291–1296. doi: 10.1093/jxb/44.8.1291. [DOI] [Google Scholar]
- Nayaka SC, Shetty HS, Satyavathi CT, Yadav RS, Kishor PK, Nagaraju M, Anoop T, Kumar MM, Kuriakose B, Chakravartty N. Draft genome sequence of Sclerospora graminicola, the pearl millet downy mildew pathogen. Biotechnol Rep. 2017;16:18–20. doi: 10.1016/j.btre.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M-A, Dow JM, Molinaro A, Parrilli M. Invited review: priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res. 2007;13(2):69–84. doi: 10.1177/0968051907079399. [DOI] [PubMed] [Google Scholar]
- Niranjan-Raj S, Lavanya S, Amruthesh K, Niranjana S, Shetty HS. Comparative evaluation of Pseudomonas fluorescens and their lipopolysaccharides as implicated in induction of resistance against pearl millet downy mildew. Arch Phytopathol Plant Protect. 2011;44(13):1285–1299. doi: 10.1080/03235408.2010.493750. [DOI] [Google Scholar]
- Noorbakhsh Z, Taheri P. Nitric oxide: a signaling molecule which activates cell wall-associated defense of tomato against Rhizoctonia solani. Eur J Plant Pathol. 2016;144(3):551–568. doi: 10.1007/s10658-015-0794-5. [DOI] [Google Scholar]
- Prakash HS, Nayaka CS, Kini KR (2014) Downy Mildew disease of pearl millet and its control. In: Future challenges in crop protection against fungal pathogens. Springer, pp 109–129
- Prockop DJ, Udenfriend S. A specific method for the analysis of hydroxyproline in tissues and urine. Anal Biochem. 1960;1:228–239. doi: 10.1016/0003-2697(60)90050-6. [DOI] [PubMed] [Google Scholar]
- Pushpalatha H, Mythrashree S, Shetty R, Geetha N, Sharathchandra R, Amruthesh K, Shetty HS. Ability of vitamins to induce downy mildew disease resistance and growth promotion in pearl millet. Crop Protection. 2007;26(11):1674–1681. doi: 10.1016/j.cropro.2007.02.012. [DOI] [Google Scholar]
- Raj SN, Lavanya S, Amruthesh K, Niranjana S, Reddy M, Shetty HS. Histo-chemical changes induced by PGPR during induction of resistance in pearl millet against downy mildew disease. Biol Control. 2012;60(2):90–102. doi: 10.1016/j.biocontrol.2011.10.011. [DOI] [Google Scholar]
- Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R. Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Protect. 2001;20(1):1–11. doi: 10.1016/S0261-2194(00)00056-9. [DOI] [Google Scholar]
- Sarosh BR, Sivaramakrishnan S, Shetty HS. Elicitation of defense related enzymes and resistance by L-methionine in pearl millet against downy mildew disease caused by Sclerospora graminicola. Plant Physiol Biochem. 2005;43(8):808–815. doi: 10.1016/j.plaphy.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Siddaiah CN, Satyanarayana NR, Mudili V, Gupta VK, Gurunathan S, Rangappa S, Huntrike SS, Srivastava RK. Elicitation of resistance and associated defense responses in Trichoderma hamatum induced protection against pearl millet downy mildew pathogen. Sci Rep. 2017;7:43991. doi: 10.1038/srep43991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddaiah CN, Prasanth KVH, Satyanarayana NR, Mudili V, Gupta VK, Kalagatur NK, Satyavati T, Dai X-F, Chen J-Y, Mocan A. Chitosan nanoparticles having higher degree of acetylation induce resistance against pearl millet downy mildew through nitric oxide generation. Sci Rep. 2018;8(1):2485. doi: 10.1038/s41598-017-19016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Molinaro A, Sturiale L, Dow JM, Erbs G, Lanzetta R, Newman M-A, Parrilli M. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J Biol Chem. 2005;280(39):33660–33668. doi: 10.1074/jbc.M506254200. [DOI] [PubMed] [Google Scholar]
- Singh S, Gopinath R. A seedling inoculation technique for detecting downy mildew resistance in pearl millet. Plant Dis. 1985;69(7):582–584. [Google Scholar]
- Smit F, Dubery IA. Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry. 1997;44(5):811–815. doi: 10.1016/S0031-9422(96)00595-X. [DOI] [Google Scholar]
- Sujeeth N, Deepak S, Shailasree S, Kini RK, Shetty SH, Hille J. Hydroxyproline-rich glycoproteins accumulate in pearl millet after seed treatment with elicitors of defense responses against Sclerospora graminicola. Physiol Mol Plant Pathol. 2010;74(3–4):230–237. doi: 10.1016/j.pmpp.2010.03.001. [DOI] [Google Scholar]
- Sun A, Li Z. Regulatory role of nitric oxide in lipopolysaccharides-triggered plant innate immunity. Plant Sign Behav. 2013;8(1):1081–1096. doi: 10.4161/psb.22554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Nie S, Xing D. Nitric oxide-mediated maintenance of redox homeostasis contributes to NPR1-dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiol. 2012;160(2):1081–1096. doi: 10.1104/pp.112.201798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur RP, Rao VP, Sharma R. Influence of dosage, storage time and temperature on efficacy of metalaxyl-treated seed for the control of pearl millet downy mildew. Eur J Plant Pathol. 2011;129(2):353–359. doi: 10.1007/s10658-010-9679-9. [DOI] [Google Scholar]
- Uzma F, Mohan CD, Hashem A, Konappa NM, Rangappa S, Kamath PV, Singh BP, Mudili V, Gupta VK, Siddaiah CN, Chowdappa S, Alqarawi AA, Abd Allah EF. Endophytic fungi-alternative sources of cytotoxic compounds: a review. Front Pharmacol. 2018;9:309. doi: 10.3389/fphar.2018.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon L, Bakker P, Pieterse C. Systemic resistance induced by rhizosphere bacteria. Ann Rev Phytopathol. 1998;36(1):453–483. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- Walters DR, Ratsep J, Havis ND. Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot. 2013;64(5):1263–1280. doi: 10.1093/jxb/ert026. [DOI] [PubMed] [Google Scholar]
- Wang J, Higgins VJ. Nitric oxide modulates H2O2-mediated defenses in the Colletotrichum coccodes–tomato interaction. Physiol Mol Plant Pathol. 2005;67(3–5):131–137. doi: 10.1016/j.pmpp.2005.11.002. [DOI] [Google Scholar]
- Wendehenne D, Durner J, Klessig DF. Nitric oxide: a new player in plant signalling and defence responses. Curr Opin Plant Biol. 2004;7(4):449–455. doi: 10.1016/j.pbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Williams R. Advances in Plant pathology. London: Academic Press; 1984. Downy mildews of tropical cereals; pp. 1–103. [Google Scholar]
- Yadav H (2014) Project coordinators review: All India coordinated project on pearl millet-49th Annual Group Meeting
- York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P (1986) Isolation and characterization of plant cell walls and cell wall components. In: Methods in enzymology, vol 118. Elsevier, pp 3–40
- Zeidler D, Zähringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA. 2004;101(44):15811–15816. doi: 10.1073/pnas.0404536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.