Figure 2.
SHIP-1 Regulates Molecular and Metabolic Hallmarks of Trained Immunity
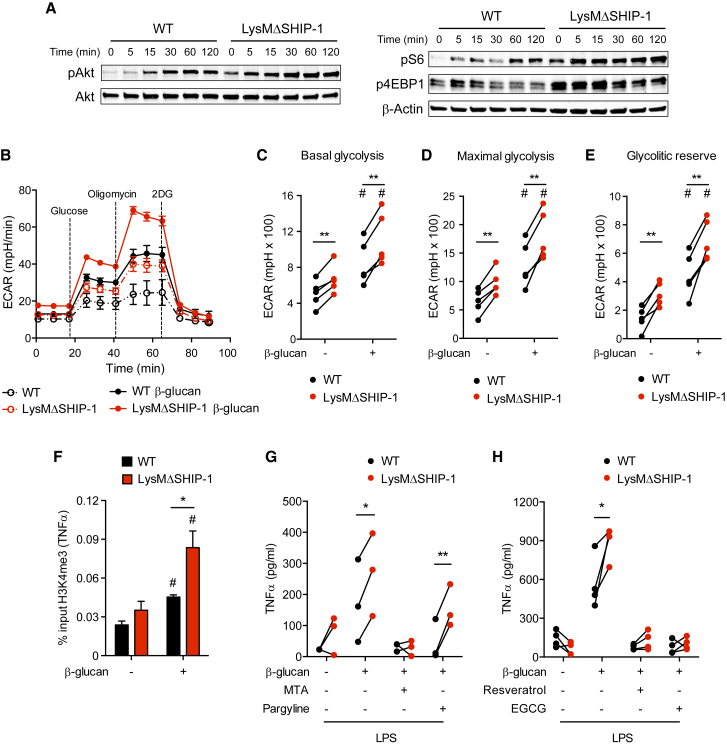
(A) BMDMs were exposed to β-glucan for the indicated time, and phospho-Akt, Akt, phospho-S6, phospho-4EBP1, and β-actin were analyzed by WB. Representative experiment of five performed. See also Figure S3.
(B–E) BMDMs were left untreated (dashed lines) or treated for 1 day with β-glucan (solid lines), washed, rested for 3 days, and re-plated in equal numbers for determination of extracellular acidification rate (ECAR) in a glycolysis stress test upon sequential addition of glucose, oligomycin, and 2-deoxyglucose (2DG) as indicated (B). Analysis of basal glycolysis (C), maximal glycolysis (D), and glycolytic reserve (E). See also Figure S4. Mean ± SEM (B) or individual data (C–E) of five independent cultures are shown.
(F) BMDMs were trained (+) or not (−) with β-glucan for 1 day, washed, and rested for 4 days and chromatin immunoprecipitation (ChIP) against H3K4me3 was performed in which enrichment on the TNFα promoter was analyzed using qPCR. Mean ± SEM of five independent experiments is shown.
(G and H) BMDMs were incubated (+) or not (−) with the methyltransferase inhibitor 5′-deoxy-5′-(methylthio)adenosine (MTA), the histone demethylase inhibitor pargyline (G), the histone deacetylase activator resveratrol, or the histone acetyltransferase inhibitor EGCG (H) for 30 min before β-glucan training and after wash-out. TNFα production was analyzed in supernatants after LPS stimulation according to Figure 1C. Individual data corresponding to three (G) or four (H) independent experiments are shown.
In (C)–(H), ∗p < 0.05 and ∗∗p < 0.01, paired Student’s t test comparing WT and LysMΔSHIP-1. In (C)–(F), #p < 0.05, paired Student’s t test comparing stimulated or not with β-glucan within the same genotype.