Fig. 1.
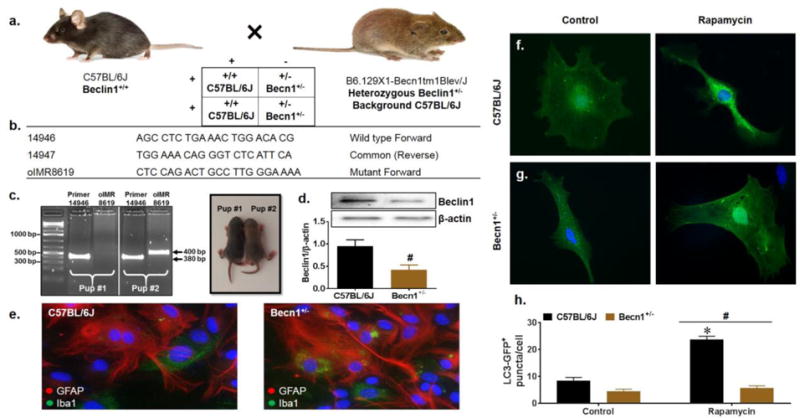
Characterization of Beclin1-deficient mixed glia. C57BL/6J (Beclin1+/+) and Becn1+/− mutant breeding scheme (a). Primers corresponding to the wild type and mutant beclin1 DNA sequence used for genotyping (b). Pups at P4-P6 were distinguished by coat color and mutation of the beclin1 gene was confirmed at the DNA and protein levels by PCR genotyping (c) and western blot (d). Lanes are grouped according to sample number (pup #1 vs pup #2) and labeled according to the primer sequence used for amplification. Wild type sequence generates a PCR product of 380 bp and mutant sequence generates a PCR product of 400bp (indicated by black arrows). Beclin1 protein expression from isolated primary glia was normalized to β-actin. Data is presented as the mean ± SEM of 4 independent experiments. Immunocytochemistry for glial cell presence identifying GFAP+ astrocytes in red and Iba1+ microglia in green (e). Representative images were acquired at 63X magnification. Representative images assessing basal autophagy induction by LC3-GFP puncta quantification upon treatment with the autophagy inducer rapamycin. Data was quantified from 3 independent experiments, averaging puncta per cell for 5 fields of view presented as mean ± SEM. P < 0.05 * vs. Control; # vs. C57BL/6J
