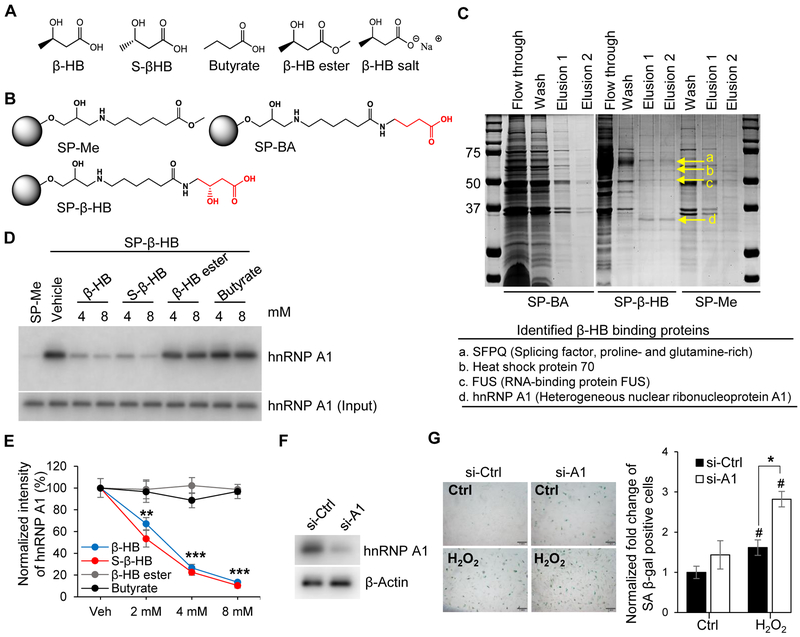
Figure 3. Proteomics approach to identify the target protein of β-HB action.
(A) Structures of β-HB and its analogues: S-β-HB, Butyrate, β-HB ester, and β-HB salt.
(B) Structures of negative control beads (SP-Me), β-HB, and butyrate-conjugated Sepharose beads (SP-β-HB and SP-BA).
(C) Representative image of affinity chromatography using SP-β-HB, SP-BA, and SP-Me. Binding proteins were eluted with β-HB gradient buffer (0.1 Μ Tris-HCl, β-HB 0.5–8 mM), then stained with Coomassie brilliant blue dye. Four proteins (SFPQ, Heat shock protein 70, FUS, and hnRNP A1) were identified by MALDI TOF MS.
(D) Representative Western blots obtained from three individual experiments. Pull-down assays using SP-Me (negative control) and SP-β-HB (positive control). Cell lysates were pretreated with competitors (β-HB, S-β-HB, β-HB ester, and Butyrate) to validate specific binding with hnRNP A1. n = 3.
(E) Quantitation of Figure 3D. **p < 0.01, ***p < 0.001 vehicle vs. treated.
(F) Representative Western blots for hnRNP A1 showing the knockdown efficiency of siRNA targeting hnRNP A1.
(G) Representative images of SA β-gal staining and quantitation of SA β-gal-positive cells in hnRNP A1-silenced HUVECs treated with or without H2O2 (150 μM). Scale bar, 1 mm. #p < 0.05 Control vs. H2O2, *p < 0.05 H2O2 + si-Ctrl vs. H2O2 + si-A1.
Data are presented as mean ± standard error of mean (SEM). NS, non-significant; si-Ctrl, control siRNA; si-A1, hnRNP A1 siRNA.