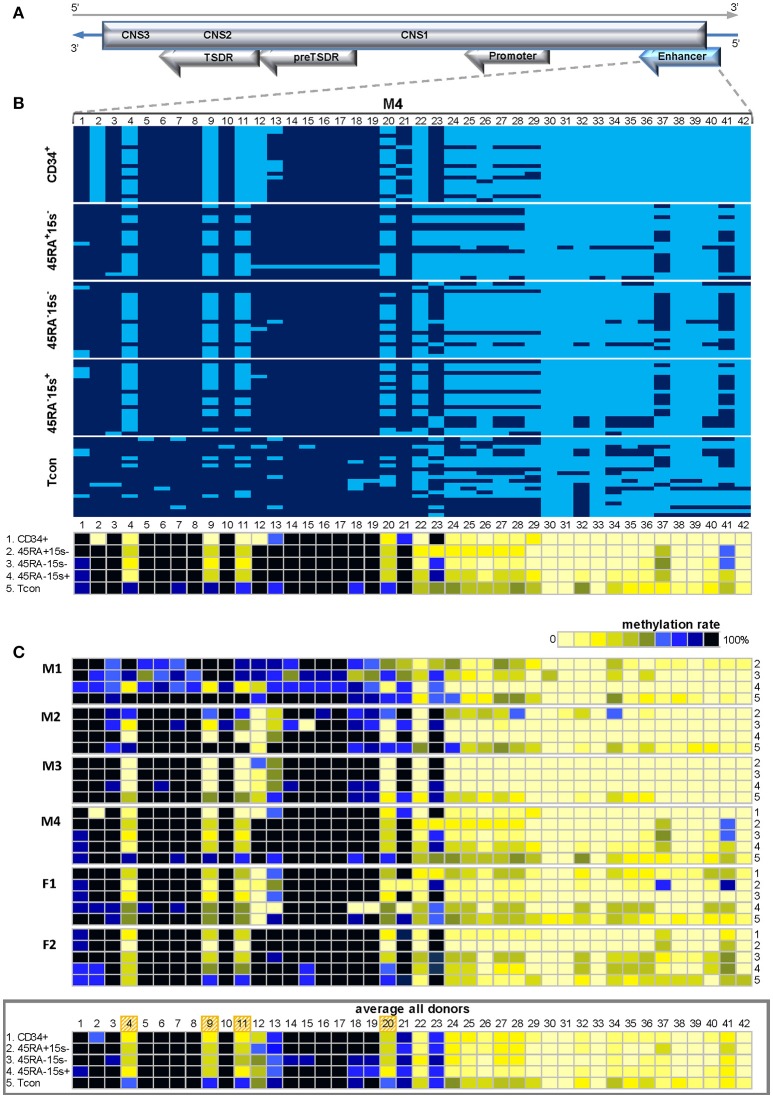
Figure 4.
Methylation pattern of FOXP3 enhancer. CpGs 4, 9, 11, 20, and 24–28 within FOXP3 enhancer are more demethylated in CD34+ cells and Treg subtypes as compared to Tcon. (A) Schematic presentation of the FOXP3 enhancer encoded in the reverse strand of Xp11.23. (B) Methylation pattern of the FOXP3 enhancer in five cell populations: CD34+ (1), CD45RA+CD15s− (2), CD45RA−CD15s− (3), CD45RA−CD15s+ (4), and Tcon (5), of representative donor M4. CpGs 1-42 are numbered relative to the 5′-3′ direction of the coding (reverse) strand. Each horizontal line represents DNA from one cell with unmodified C in light blue and mC in dark blue. Methylation percentage for each CpG site was calculated based on the number of mCs in a total of 20 and summarized in panels below. (C) Methylation patterns of the FOXP3 enhancer of individual donors with the average methylation pattern for all donors presented in the panel below. See also Supplementary Figures 1, 4.