Figure 2.
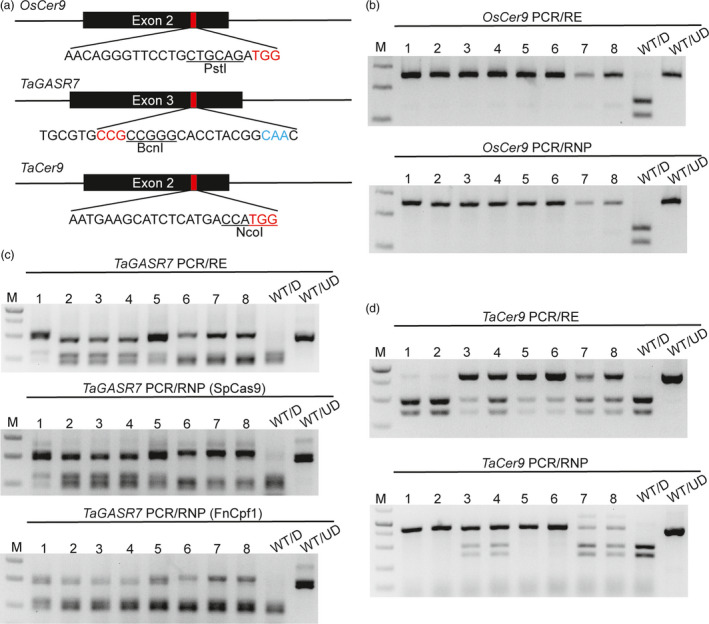
Genotyping of mutants induced by CRISPR/Cas9 reagents using PCR/RNP and PCR/RE in plants. (a) Three sgRNAs were designed to target rice OsCer9 and wheat TaGASR7 and TaCer9, respectively. The PAM sequences are highlighted in red and the corresponding restriction enzyme sites are underlined and labelled. The PAM sequence of FnCpf1 (5′‐CAA‐3′) in the sg‐TaGASR7 is highlighted in blue. (b) The results of PCR/RE and PCR/RNP assays to detect eight homozygous oscer9 mutants induced by CRISPR/Cas9. (c) The results of PCR/RE and PCR/RNP (SpCas9 and FnCpf1) assays to detect eight representative mutants induced by gasr7‐IVTs. (d) Agarose gel analysis of eight CRISPR/Cas9‐induced tacer9 mutants detected by PCR/RE and PCR/RNP. ‘WT/D’: wild‐type amplicons digested with T7EI, restriction enzyme or CRISPR/Cas9 RNP; ‘WT/UD’: wild‐type amplicon not digested.
