Figure 5.
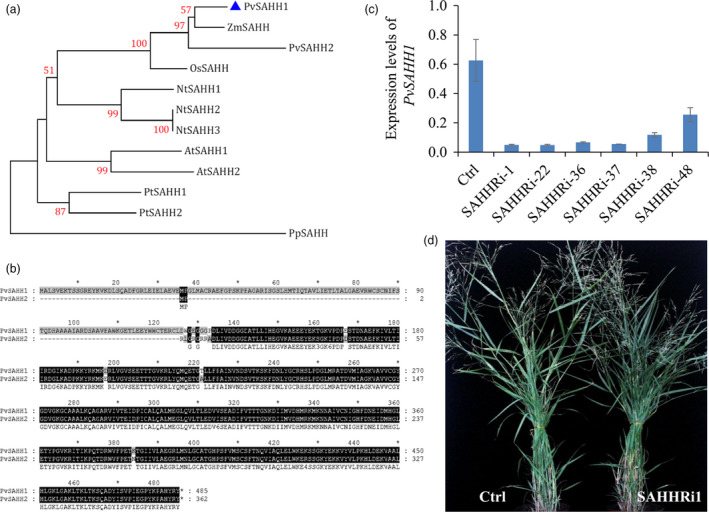
Characterization of SAHH‐RNAi transgenic switchgrass plants. (a) Phylogenetic tree of the SAHH orthologs in monocot and dicot plant species. A maximum likelihood tree was constructed in PhyML version 3.0 on the basis of multiple alignments of the deduce protein sequences from Panicum virgatum (PvSAHH1, Pavir.1NG430900; PvSAHH2, Pavir.6KG019200), Zea mays (ZmSAHH, GRMZM2G015295), Oryza sativa (OsSAHH, LOC_Os11g26850), Arabidopsis thaliana (AtSAHH1, AT4G13940.1; AtSAHH2, AT3G23810.1), Nicotiana tobacum (NtSAHH1, mRNA_62654_cds; NtSAHH2, mRNA_112781_cds; NtSAHH3, mRNA_49476_cds), Populus trichocarpa (PtSAHH1, POPTR_0017s08610.1; PtSAHH2, POPTR_0001s32780.1) and Physcomitrella patens (Pp1s20_229V6.1). Bootstrap values (>50%) based on 1000 replications are indicated at nodes. The above sequence data are retrieved from Phytozome and Sol Genomics Network. (b) Sequence alignment between PvSAHH1 and PvSAHH2 identified from switchgrass genome database. (c) Quantitative real‐time PCR analysis of PvSAHH1 transcript abundance in the SAHH‐RNAi transgenic lines. Switchgrass UBQ was used as the reference for normalization. Stems at E4 stage were collected. Value are mean ± SE (n = 3). (d) Morphological characterization of transgenic switchgrass plants with down‐regulation of PvSAHH1. The control plants (Ctrl) for SAHH‐RNAi transgenic lines were generated with pANIC8B empty vector.
