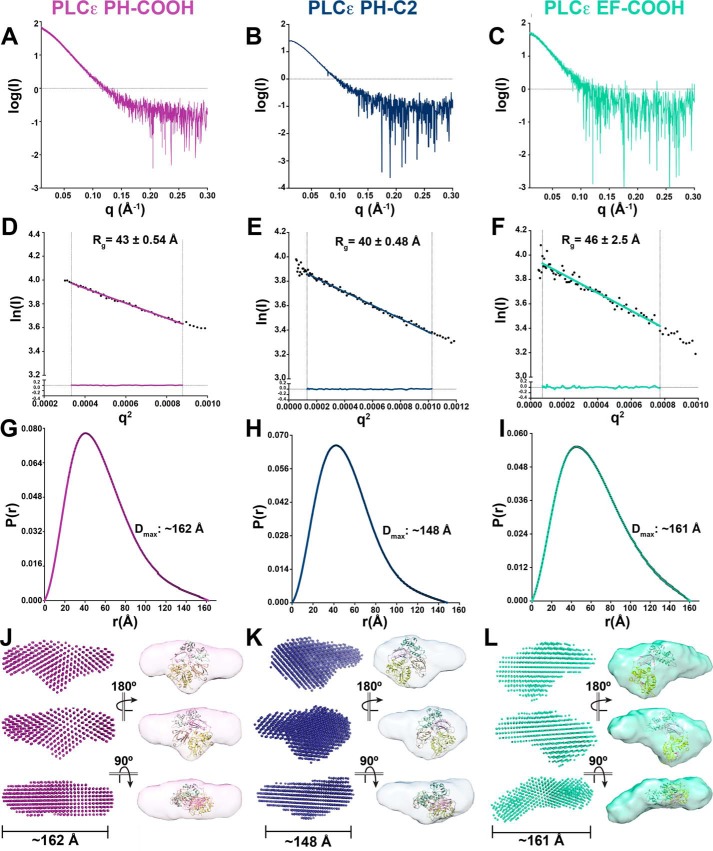
Figure 2.
SAXS envelopes reveal the solution architectures of PLCϵ variants. A–C, raw scattering curves for PLCϵ PH-COOH (A), PH-C2 (B), and EF-COOH (C). D–F, Guinier analyses of low q values, ln(I) (beam intensity) versus q2 (scattering angle) with Rg of PH-COOH, PH-C2, and EF-COOH, respectively. Fitting of the linear regressions to the data is represented by residuals, shown at the bottom of the plots, demonstrating that the proteins are monomeric in solution. G-I, pair distance distribution functions (P(r)) indicating elongated envelopes for PH-COOH, PH-C2, and EF-COOH variants, respectively. Estimated maximum intramolecular distances (Dmax) are provided. Ab initio envelope models (left) and equivalent envelopes rendered as volumes (right) show protrusions in PH-COOH (J) and PH-C2 (K) and lack of density, likely corresponding to the missing RA domains in PH-C2. As a reference, the crystal structure of the PLCβ3 core (colored as in Fig. 1A; PDB entry 3OHM (12)) is fit within the SAXS-derived envelopes such that the PH domain is oriented toward the extended protrusion and the C2 domain toward the additional density on the opposite side of the envelope. In L (EF-COOH), the domain is extended similarly to PH-COOH, but electron density is lost from the protrusion and the center of the molecule due to loss of the PH domain. The PLCβ3 core lacking the PH domain is fit within the density as described for J and K.