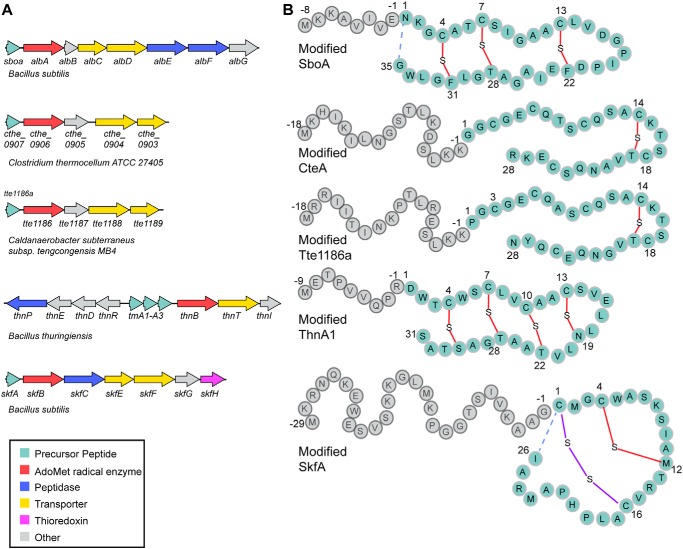
Figure 1.
Sactipeptide biosynthesis. A, gene scheme of several RiPP biosynthetic gene clusters involved in the biosynthesis of sactipeptides. These biosynthetic clusters encode precursor peptides (cyan); AdoMet radical enzymes (red), which are responsible for installing the unique sactionine linkages characteristic of the sactipeptide natural products; peptidases (blue), which are responsible for cleaving the leader peptide; and ABC transporters, which are involved in transporting the sactipeptide out of the producing organism. B, the precursor peptides for each biosynthetic gene cluster in A are shown with the leader peptide sequence, which binds to the biosynthetic enzymes, in gray and the mature peptide, which undergoes post-translational modifications, in cyan. The sactionine linkages formed between the sulfur atom of a cysteine residue and the α-carbon of an acceptor residue are shown in red. Final maturation steps of SboA and SkfA involve an N- to C-terminal macrocyclization (dashed blue lines) step concurrent with a leader peptide cleavage step. Maturation of SkfA also includes disulfide bond formation (purple).