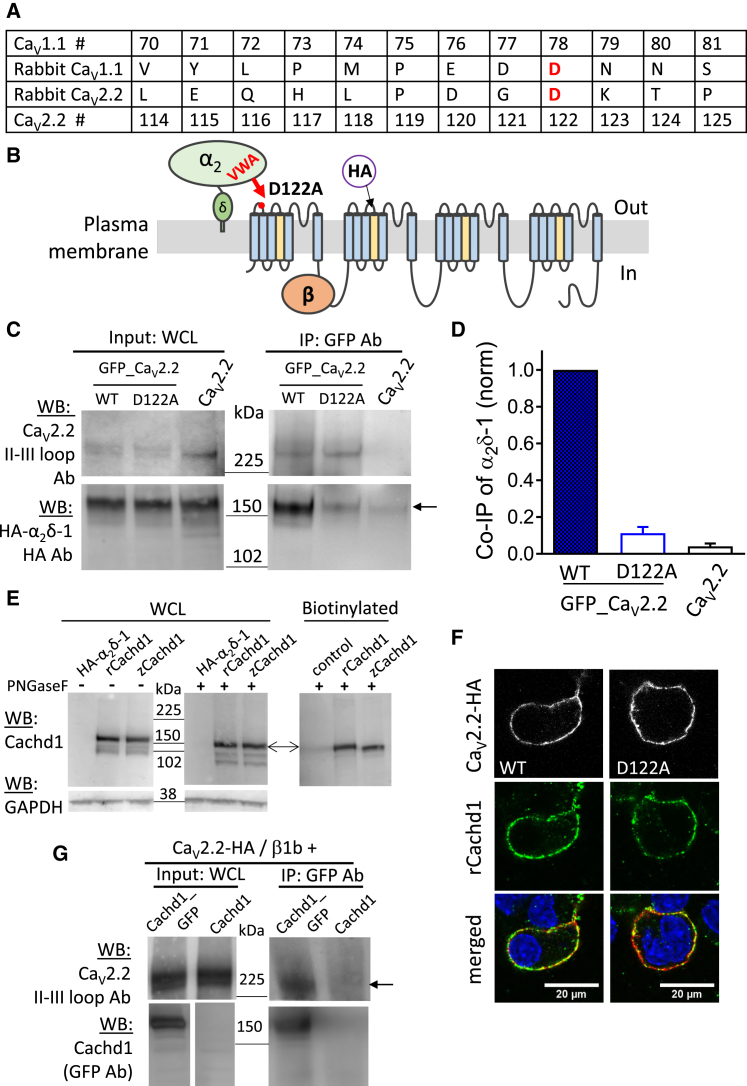
Figure 1.
Effect of D122A Mutation in CaV2.2 on Interaction with α2δ-1 and Cachd1
(A) Sequence alignment of the VWA domain interaction site on CaV1.1 in comparison with the rabbit CaV2.2 used in this study, showing the position of D122 in the first extracellular loop of CaV2.2. Residue numbering is shown (#).
(B) Diagram of the putative CaV2.2 interaction site with the VWA domain of α2δ-1, showing the position of the D122A mutation and the HA epitope tag.
(C) IP of GFP_CaV2.2, and co-immunoprecipitation (coIP) of α2δ-1. WCL input (left) and IP (right) for WT and D122A mutant GFP_CaV2.2 and untagged CaV2.2 control (top) and for HA-tagged α2δ-1 (bottom). IP was performed with GFP Ab and pulled down both WT and D122A GFP_CaV2.2 (top right). CoIP of HA-tagged α2δ-1 is shown in at the bottom right (arrow).
(D) Quantification of coIP of α2δ-1 with WT GFP_CaV2.2 (solid blue bar) compared with D122A GFP_CaV2.2 (open blue bar) and control CaV2.2 (open black bar); mean ± SEM of 5 experiments.
(E) Western blot using Cachd1 Ab of WCL from tsA-201 cells transfected with α2δ-1 as a control (lane 1), rCachd1 (lane 2), and zCachd1 (lane 3). Left: prior to deglycosylation with PNGase F. Center: after deglycosylation. Bottom: glyceraldehyde 3-phosphate dehydrogenase (GAPDH) loading control. Right: a separate experiment after cell surface biotinylation and deglycosylation; the control here was untransfected cells. The arrow indicates a major Cachd1 band.
(F) Representative confocal images of N2A cells expressing CaV2.2 HA WT (left) or D122A (right) with β1b and rCachd1. Cells were not permeabilized and incubated with rat anti-HA and rabbit anti-Cachd1 Abs for 1 hr to show extracellular HA staining on the plasma membrane (top row, white) and Cachd1 (center row, green). Merged images (with HA in red and co-localization in yellow) are shown at the bottom; DAPI was used to stain the nuclei (blue). Scale bars, 20 μm.
(G) IP of Cachd1_GFP and coIP of CaV2.2. Shown are WCL input (left) and IP (right). Top: CaV2.2. Bottom: zCachd1_GFP (left lane) and untagged zCachd1 (right lane; both lanes are from the same blot). IP was performed with GFP Ab and pulled down both Cachd1_GFP (bottom) and CaV2.2 (top right, arrow). Lack of coIP of CaV2.2 with untagged Cachd1 is shown in the right lane. Data are representative of n = 6 experiments. zCachd1 expression in WCL is confirmed in Figure S1C.