Figure 1.
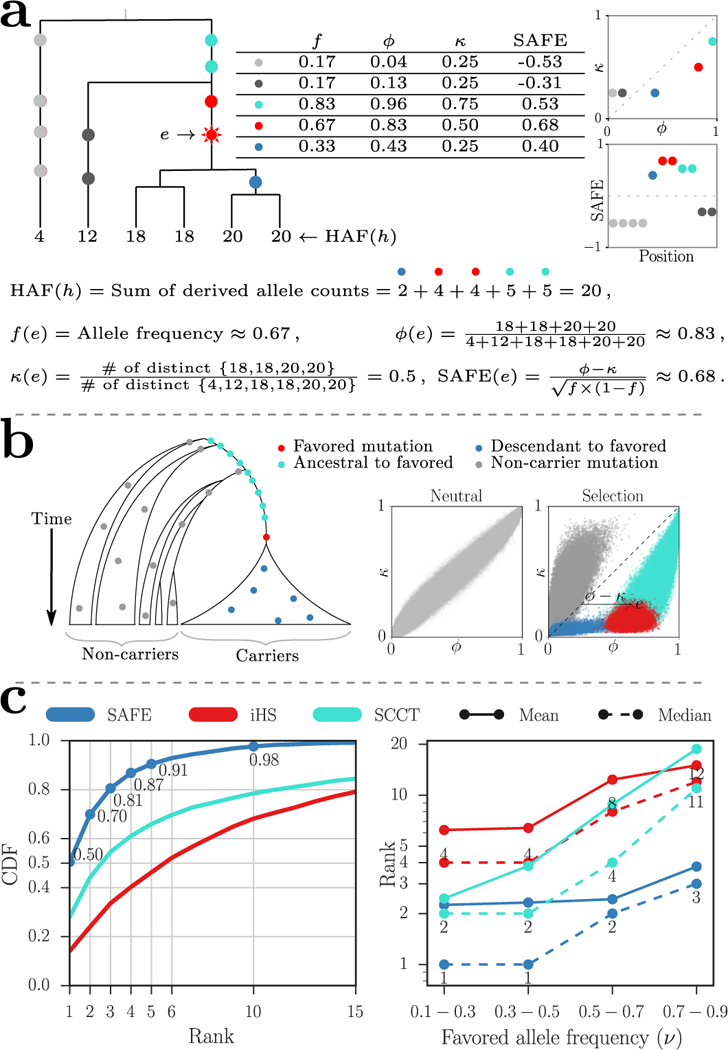
Illustration and performance of the SAFE method. (a) The HAF score for haplotype h is the sum of the derived allele counts of the mutations on h. Carriers of the favored mutation have higher fraction of the total HAF score of the sample (high ϕ)6, and lower number of distinct haplotypes compared to non-carriers (low κ). (b) Schematic of a no-recombination (for exposition purposes) genealogy under a selective sweep. The mutations can be categorized as ‘non-carrier’ (gray), ‘ancestral to favored’ (turquoise) arising prior to the favored mutation, and ‘descendant to favored’ (blue) that arise on haplotypes carrying the favored mutations but after the favored mutation, and the favored mutation itself (red). In the right panels, simulations showing ϕ versus κ values for each variant in a neutral evolution and a selective sweep for 1000 simulations with favored allele frequency (ν0 = 0.5) and default values for other simulation parameters (see Online Methods). The joint-distribution of ϕ and κ, in a selective sweep, changes in a dramatic but predictable manner that separates out non-carrier (gray), descendant (blue), and ancestral (turquoise) mutations from the favored (red) mutations. The SAFE score computes a normalized difference of the two statistics, ϕ and κ. (c) Performance (favored mutation rank) of SAFE compared to iHS and SCCT on 50 kbp windows with 1000 simulations per frequency bin. The simulations were performed with default parameter values (see Online Methods) for a fixed population size with ongoing selective sweeps. The left panel combines all allele frequencies while the right panel shows median and mean ranks for replicates divided into four bins.
