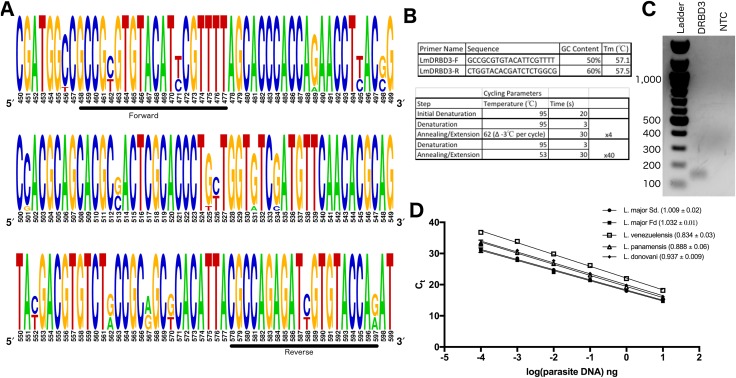
Figure 1. Primer design and optimization of DRBD3 based qPCR for parasite quantification.
(A) Multisequence alignment based on 11 homologous sequences to L. major Sd. DRBD3 found using NCBI Blast. (B) Primer sequences and cycling parameters used. (C) DRBD3 primers amplify a 140 bp product specifically. Product visualized with ethidium bromide staining of a 1% agarose gel run and compared to New England BioLabs 100 bp ladder. (D) DRBD3 primers amplify diverse Leishmania spp. A representative plot of Ct value vs log dilution of parasite burden is shown. The average primer efficiency (±standard error of the mean) is indicated in parentheses for the following species: L. major Seidman (n = 6), L. major Friedlin (n = 5), L. venezuelensis (n = 5), L. panamensis (n = 3), and L. donovani (n = 3).