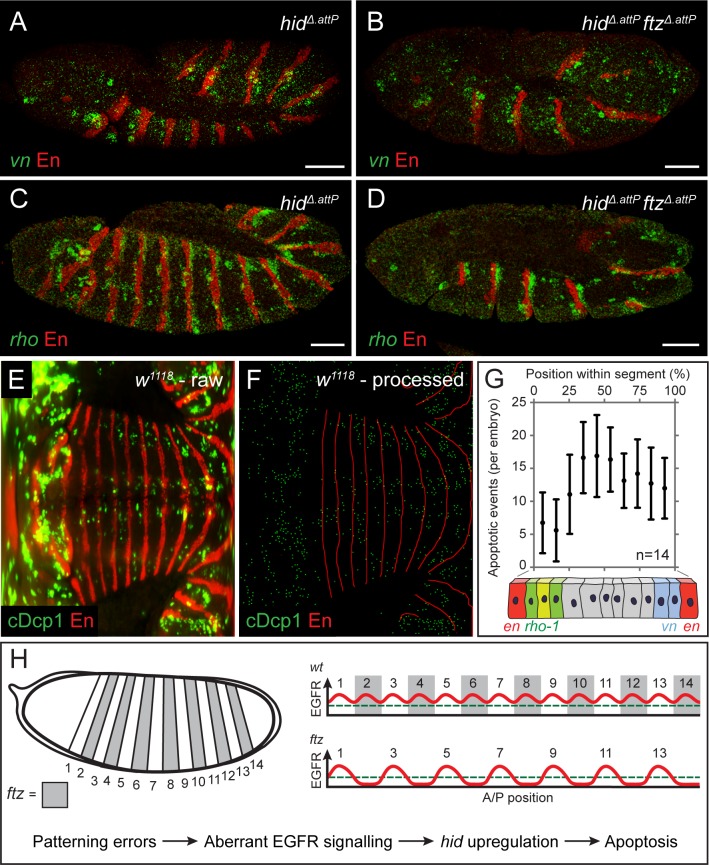
Fig 4. Patterned expression of vn and rho-1 ensure epidermal cell survival.
(A, B) Expression of vn (green) in control hidΔ.attP (A) and hidΔ.attP ftzΔ.attP (B) stage 12 homozygotes, as detected by fluorescent in situ hybridisation. hidΔ.attP is included in the background to avoid any confounding effects of apoptosis. (C, D) Expression of rho-1 (green) in hidΔ.attP (C) and hidΔ.attP ftzΔ.attP (D) stage 12 homozygotes, as detected by fluorescent in situ hybridisation. Scale bars 50 μm. (E) Representative raw mSPIM image of a stage 12 w1118 embryo (control) stained with anti-cDcp1 and anti-En (red). Anterior is to the left and posterior to the right, with the ventral midline running horizontally through the middle. (F) Processed version of the image shown in E. cDcp1-positive cells were segmented and reduced to individual pixels (green). Similarly, stripes of en expression were reduced to a skeleton (red) to allow the position of each apoptotic cell to be mapped along the A/P axis of the segment and thus to derive a relative position applicable to all segments. (G) Histogram displaying the frequency of apoptosis along the A/P axis of the segment. Illustration of the en, rho-1, and vn expression domains is included to indicate the approximate sites of ligand production. Error bars show standard deviation. (H) Model explaining the pattern of apoptosis in ftz and other segmentation mutants. In normal development, the segmental expression of EGFR ligands maintains signaling activity above the threshold level required for cell survival (green dashed line). When patterning errors disrupt the landscape of EGFR activity, signaling falls below the survival threshold, triggering hid up-regulation and patterned apoptosis. The underlying data for (G) can be found in S1 Data. A/P, anterior/posterior; cDcp1, cleaved Death caspase-1; EGFR, epidermal growth factor receptor; en, engrailed; ftz, fushi-tarazu; hid, head involution defective; mSPIM, multiview single plane illumination microscopy; rho-1, rhomboid; vn, vein; w1118, white1118.