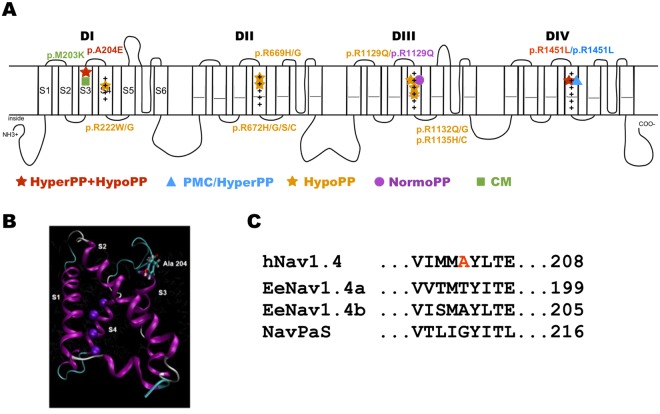
Figure 1.
Schema of the pore-forming α subunit of Nav1.4 and location of A204E. (A) The pore-forming α subunit of hNav1.4 is composed of 1.836 amino acid residues forming 4 homologous domains (DI-DIV). Each domain is composed of 6 transmembrane segments (S1–S6). The S1–S4 segments of each domain form the voltage-sensor domain, with the positively-charged S4 segments acting as voltage-sensors while the S5 and S6 segments form the selective α pore. More than 70 mutations have been described in human Nav1.4. Only the ones located in DIS3 or phenotypically-related to A204E are listed on the schema: p.M203K located in DIS3 and associated to a congenital myopathy phenotype; the hypoPP missense mutations substituting positively-charged (+) residues in S4 segments; p.R1451L resulting in a PP phenotype combining hyper- and hypo-PP. The p.R1129Q and p.R1451L are associated with two distinct phenotypes: hypoPP or normoPP (R1129Q), and paramyotonia congenita with hyperPP or PP combining hyper- and hypo-PP signs (R1451L). Hyper PP = hyperkalaemic periodic paralysis; hypoPP = hypokalaemic periodic paralysis; PMC = paramyotonia congenita; NormoPP = normokalaemic PP; CM = congenital myopathy. (B) Model of voltage-sensor domain I based on NavAb crystal structure40 showing the position of the Ala204 residue on the extracellular side of S3. (C) Alignment of the hNav1.4 amino acid sequence around the A204 residue with eukaryotic EeNav1.4 (electric eel) and NaVPaS (putative Nav channel from the American cockroach) channels whose cryo-electron microscopy structure was determined with a high-level resolution. The A204 residue is not conserved in these orthologs except in EeNav1.4b.