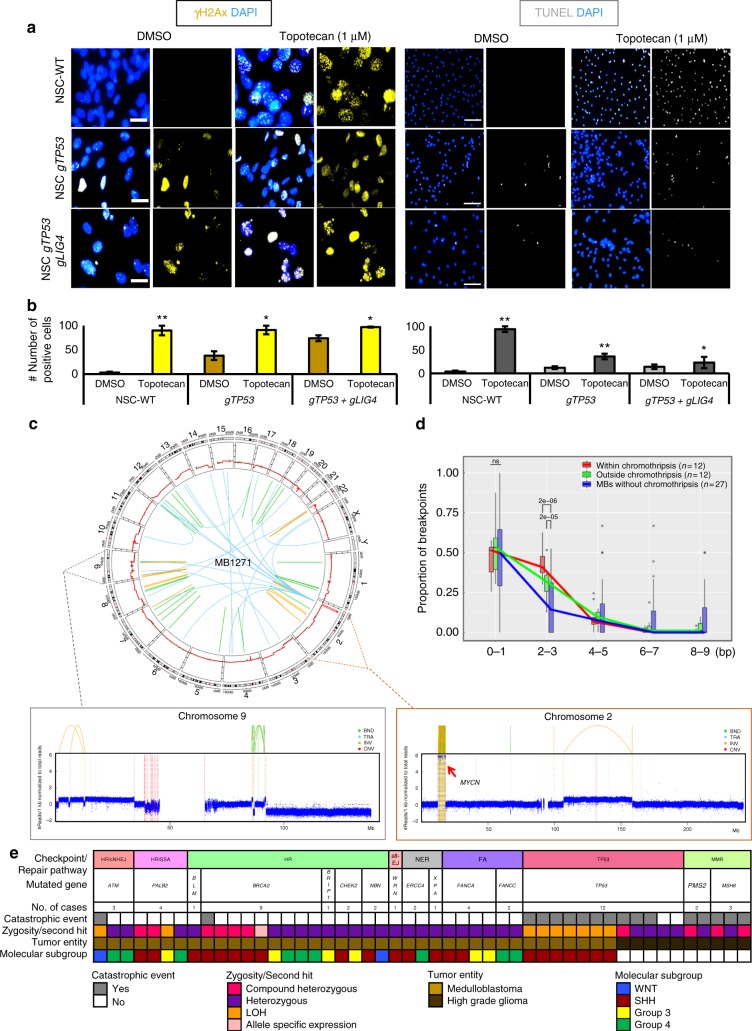
Fig. 5.
Analysis of human neural progenitor cells, medulloblastomas and high-grade gliomas with DNA repair defects. a Response to DNA damage after CRISPR/Cas9-mediated disruption of TP53 and LIG4 in human iPSC-derived neural progenitor cells. Left panel, immunofluorescence analysis of DSB marker gH2AX. Right panel, TUNEL. Topotecan treatment (1 µM) was applied for 48 h to induce DNA DSBs. b Quantifications of the numbers of positive cells in topotecan-treated and control cells after disruption of TP53 and LIG4. Average values and standard deviations for three independent experiments are shown. Unpaired t-tests were used to test for statistical significance. c Chromothripsis in a human medulloblastoma with BRCA2 deficiency. Circos plot and coverage plots showing rearrangements associated with chromothripsis on chromosome 9 and MYCN amplification on chromosome 2. d Analysis of the length of the microhomologies in human SHH MBs at the breakpoints due to catastrophic events (red), on chromosome regions not affected by catastrophic events in the same tumors (green) or genome-wide in MBs not showing chromothripsis (blue). The median for each boxplot is indicated by the center line, the rectangle span the interquartile range (whiskers show maximum and minimum values). e Heatmap showing complex genome rearrangements in human medulloblastomas (n = 32) and high-grade gliomas (n = 17) with pathogenic germline mutations in major components of DNA repair and checkpoint pathways (re-analysis of whole-genome sequencing data from two published studies14,26)