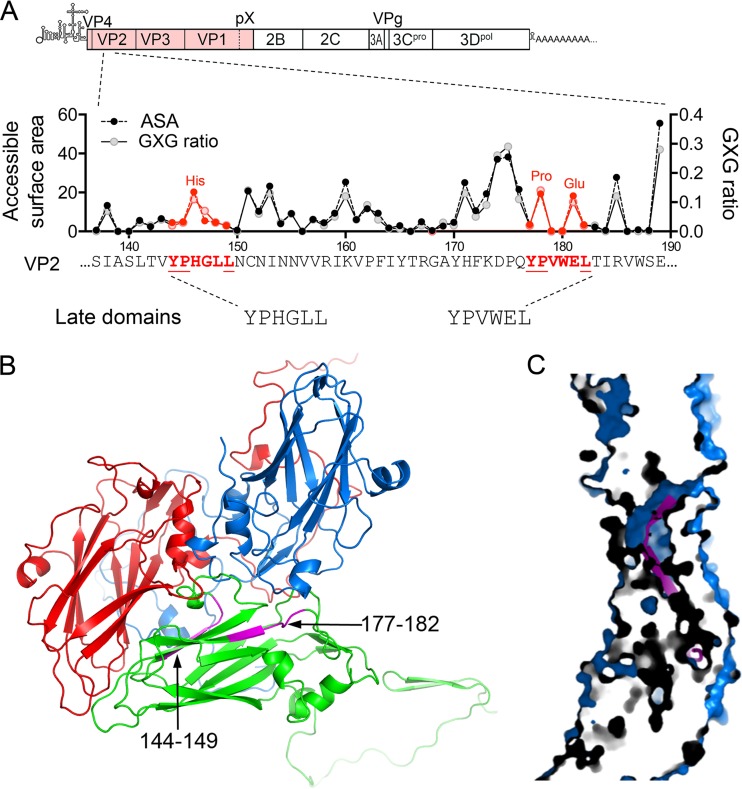
FIG 1.
HAV VP2 YPX3L late domain motifs. (A) Schematic representation of the two tandem late domain motifs in the HAV VP2 capsid protein. The organization of the HAV genome is shown at the top with the polyprotein coding region displayed as an extended box. Below is shown a segment of the VP2 protein with the two conserved YPX3L motifs in red font and with the accessible surface area (ASA) of each residue plotted above along with the ratio to calculated GXG value (surface area of the residue relative to that in a peptide in which it is flanked on each side by Gly) based on the X-ray model of the naked HAV capsid (17). (B) Protomer subunit of the HAV capsid showing the adjacent, antiparallel orientation of the two late domain motifs (L1, residues 144 to 149, and L2, residues 177 to 182, highlighted in magenta) within the VP2 (green) β-barrel (17). VP1 is shown in blue, and VP3 is shown in red. (C) Section through the HAV capsid wall, showing the buried position of residues within the late domain motifs (magenta).