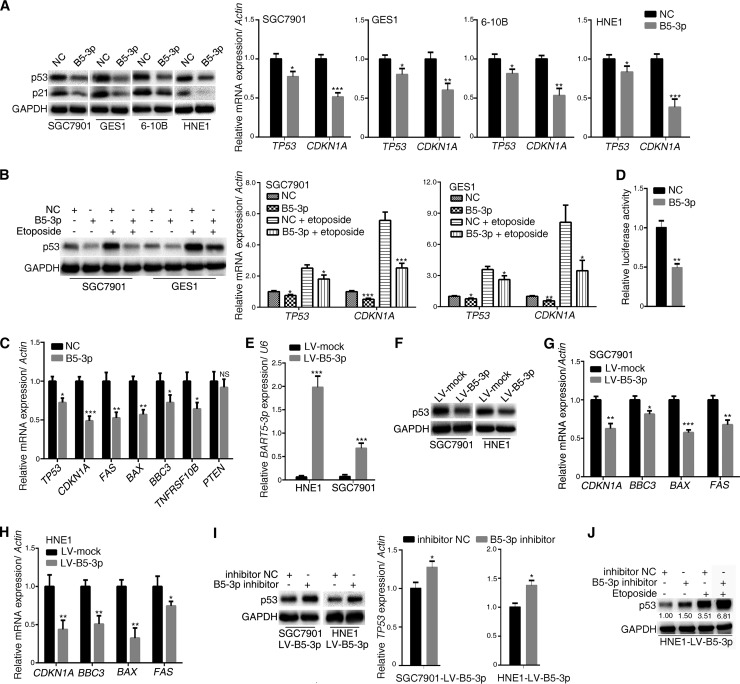
FIG 1.
BART5-3p inhibits the expression and transcriptional activity of p53 in multiple cell lines. (A) SGC7901, GES1, 6-10B, and HNE1 cells were transfected with either negative-control mimics (NC) or BART5-3p mimics (B5-3p) for 36 h. The protein levels of p53 and p21 were determined by Western blotting, and the mRNA levels of TP53 and CDKN1A were analyzed by qPCR. (B) SGC7901 and GES1 cells were transfected with either NC mimics or BART5-3p mimics for 12 h initially and then treated (or not treated) with 10 μM etoposide for another 24 h; the protein levels of p53 were determined by Western blotting, and the mRNA levels of TP53 and CDKN1A were analyzed by qPCR. (C) SGC7901 cells were transfected with NC or BART5-3p mimics for 24 h. The mRNA expression levels of the indicated genes were analyzed by qPCR. (D) SGC7901 cells were cotransfected with mimics and firefly luciferase pp53-TA-luc reporter plasmid along with Renilla luciferase reporter plasmid for 24 h before the luciferase reporter assay was performed. The data are shown as the relative firefly luciferase activity normalized to the value of Renilla luciferase. (E to H) Two stable BART5-3p-expressing cell lines were constructed by lentivirus-mediated transduction in HNE1 and SGC7901 cells. The transcriptional levels of BART5-3p were assayed by qPCR (E), and the p53 protein levels were determined by Western blotting (F); mRNA expression levels of the indicated genes were analyzed by qPCR (G, H). (I) HNE1-LV-B5-3p and SGC7901-LV-B5-3p cells were transfected with NC inhibitors or BART5-3p inhibitors for 36 h. The p53 protein levels were determined by Western blotting, and the mRNA levels of TP53 were analyzed by qPCR. (J) HNE1-LV-B5-3p cells were transfected with either NC inhibitor or BART5-3p inhibitor for 12 h initially and then treated (or not treated) with 10 μM etoposide for another 24 h. The protein expression levels of p53 were determined by Western blotting. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as protein loading control. Actin was used for normalizing the expression of mRNA. RPU6B (U6) was used for normalizing the expression of BART5-3p. Both the mean mRNA expression and mean luciferase activity were calculated from 3 independent experiments, and data are shown as the means ± standard errors of the means (SEM). NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with the control group (NC or mock).