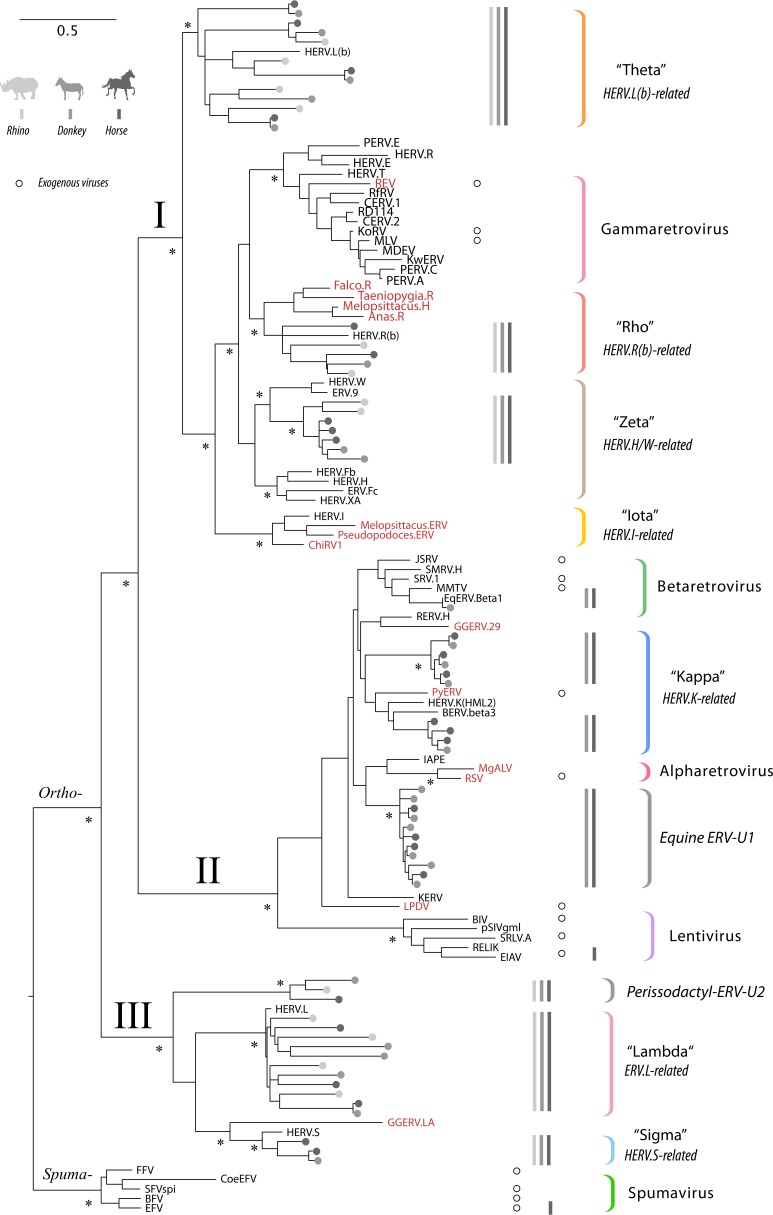
FIG 1.
Evolutionary relationships between perissodactyl endogenous, previously characterized ERVs, and exogenous retroviruses. The figure shows a maximum likelihood phylogeny reconstructed from an alignment of retroviral reverse transcriptase (RT) peptide sequences. Sequences extracted from horse, donkey, and rhino genomes are indicated by gray circles, following the color key shown in the top left corner of the figure. For previously characterized ERVs and exogenous retroviruses, taxon labels show the abbreviated names (see Table S8 in the supplemental material for complete details). Sequences derived from exogenous virus references are marked by open circles aligned with taxon labels. Sequences identified in nonmammalian hosts are indicated by red font. Retrovirus subfamilies and orthoretroviral clades (I, II, and III) are indicated on basal branches. Established retroviral genera and ERV lineages defined in this study are indicated by colored brackets. For each of these groups, the presence of sequences in the rhinoceros, donkey, and horse in each genus is indicated by gray bars, following the color key (top left corner of figure). Nodes with bootstrap support of ≥70% are indicated by an asterisk. The bar shows evolutionary distance in substitutions per site.