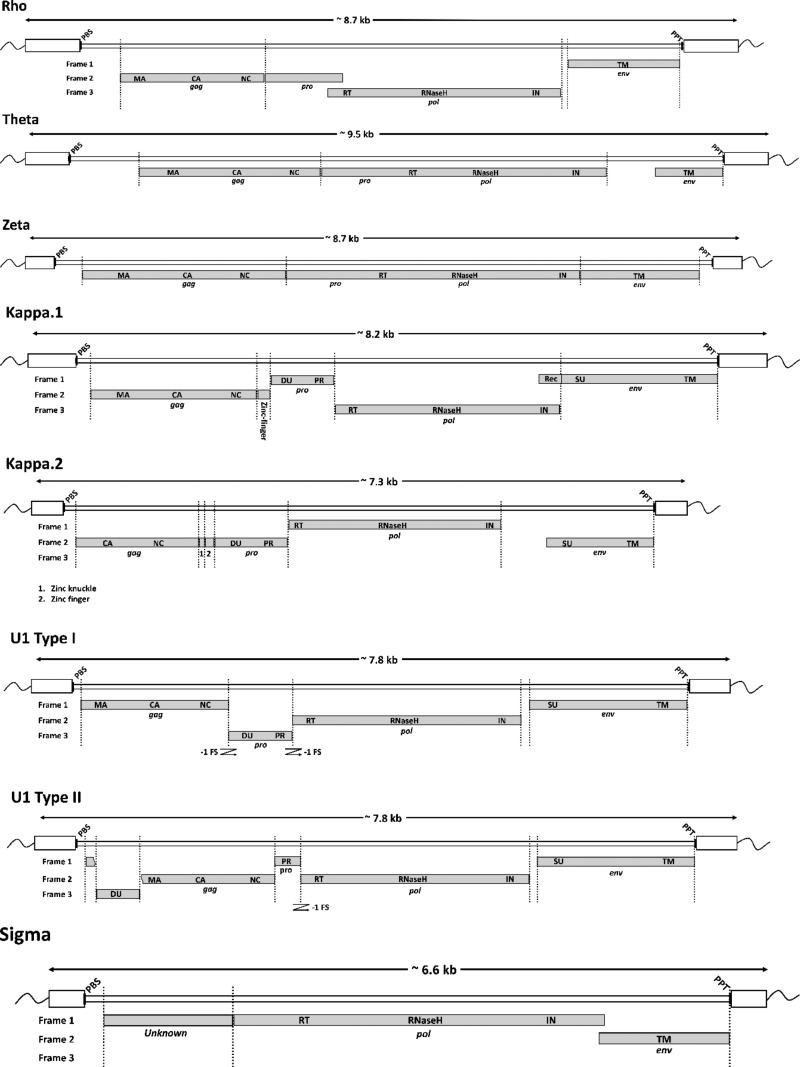
FIG 3.
Schematic representation of proviruses. The putative locations of gag-, pro-, pol-, and env-coding domains within consensus proviral genomes are indicated by gray boxes. Long terminal repeat (LTR) sequences are shown as white boxes. The estimated positions of primer binding sequence (PBS) and polypurine tract (PPT) sequences are indicated by black bars. A bar indicating the length in kilobases is shown above each genome diagram. Abbreviations: PBS, primer binding site; MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease; DU, dUTPase; RT, reverse transcriptase; IN, integrase; SU, surface glycoprotein; TM, transmembrane domain; PPT, polypurine tract.