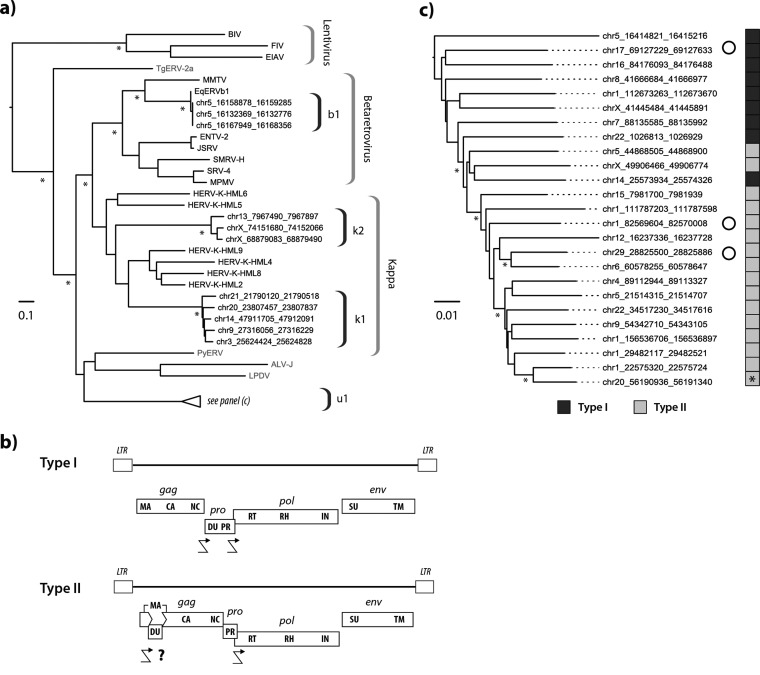
FIG 5.
Characteristics of modern equine ERVs. (a) Maximum likelihood phylogeny representing the estimated evolutionary relationships between Pol sequences derived from clade II ERVs in perissodactyl genomes and those of previously characterized ERVs and exogenous retroviruses. Taxon labels for RT sequences detected in this study indicate the species in which they were identified. Other taxon labels show the abbreviated name of the virus or ERV. Sequences identified in nonmammalian hosts are indicated in gray. Brackets on the right indicate ERV lineages and retroviral genera. Nodes with bootstrap support above 70% are indicated by asterisks. The bar shows evolutionary distance in substitutions per site. Details of taxa are provided in Table S8. chr5, chromosome 5. (b) Consensus genome structures of EqERV.U1 proviruses. Viral coding domains are shown as dark gray bars. Long terminal repeats (LTRs) are shown as boxes. Crooked arrows indicate where we have inferred translational frameshifting. For type II proviruses, we show a putative frameshift site (indicated with a question mark) that would allow expression of a matrix-dUTPase fusion protein. Abbreviations: LTR, long terminal repeat; MA, matrix; CA, capsid; NC, nucleocapsid; DU, dUTPase; PR, protease; RT, reverse transcriptase; IN, integrase; SU, surface; TM, transmembrane. (c) Maximum likelihood phylogeny of EqERV.U1 loci based on the aligned nucleotide sequences of 25 full-length proviruses. The sidebar boxes to the right of the taxa indicate the type of genome found in the element (see panel b) as indicated in the key below the tree. An asterisk on the sidebar shows the youngest provirus based on the paired LTR dating. Open circles indicate loci that show evidence of transcription based on analysis of transcriptomic data sets. Asterisks indicate nodes with bootstrap support above 70%. The bar shows evolutionary distance in substitutions per site.