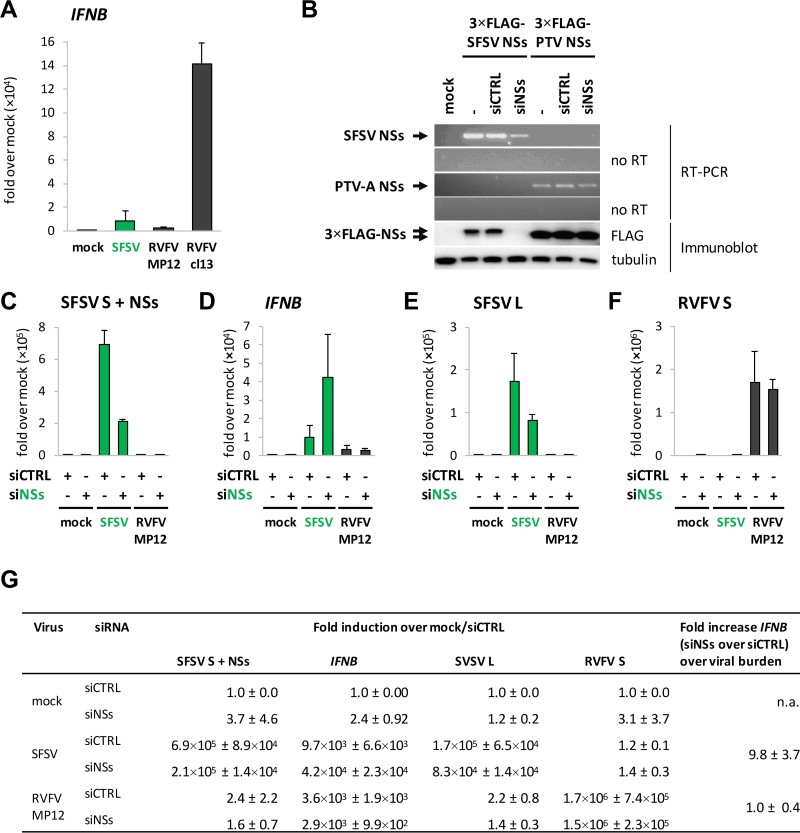
FIG 1.
SFSV NSs and IFNB induction. (A) A549 cells were infected with SFSV, RVFV MP12, or clone 13 (Cl13) at an MOI of 1, harvested 12 hpi, and analyzed by RT-qPCR analysis for IFNB (n = 4; mean ± SD). (B) A549 cells were cotransfected with expression constructs for 3×FLAG-tagged SFSV or PTV-A NSs and nontargeting control siRNA or SFSV NSs-specific siRNA. Samples were subjected to RT-PCR analysis (upper panels) and immunoblotting using anti-FLAG and anti-tubulin antibodies (lower panel) 24 h after transfection. To exclude amplification of NSs sequences from plasmid DNA, a duplicate set of reactions was performed without the reverse transcription step (no RT). (C to F) A549 cells were pretransfected with control or SFSV NSs-targeting siRNA and infected with SFSV or RVFV MP12 at an MOI of 1. RNA was isolated 12 hpi for RT-qPCR analysis for NSs-containing RNA (C), IFNB (D), the L segment of SFSV (E), and the S segment of RVFV MP12 (n = 3; means ± SD) (F). (G) Summary of the relative fold induction data depicted in panels C to F, normalized to the mock sample pretreated with control siRNA as well as the fold induction of IFNB in siNSs-treated cells over siCTRL-treated cells that occurred in a manner independent of the viral burden (means ± SD). n.a., not applicable.