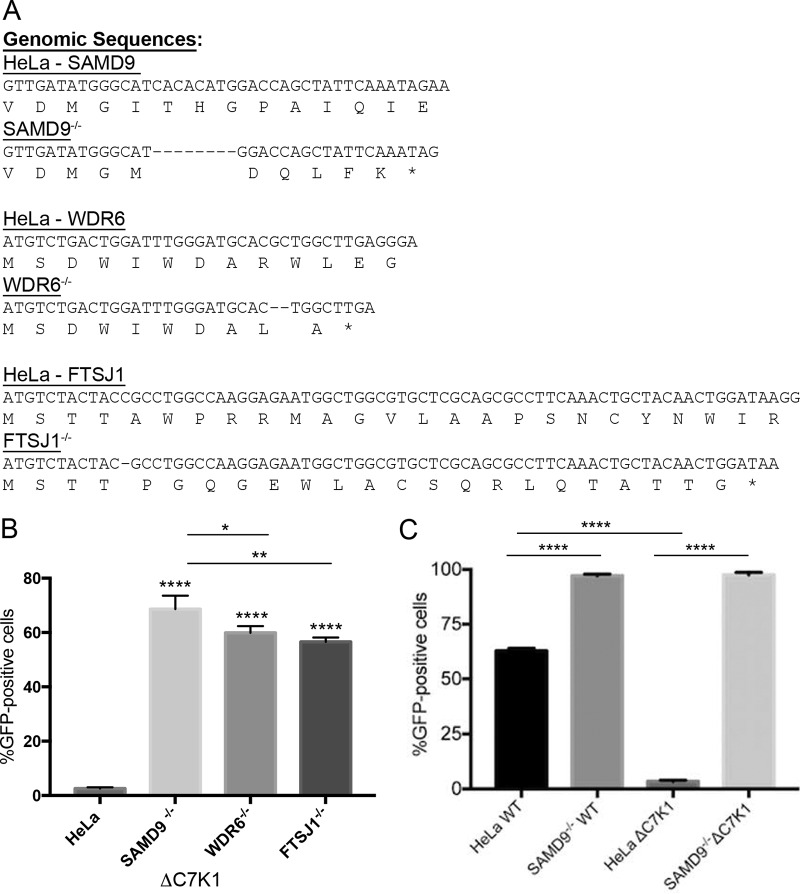
FIG 1.
Comparison of ΔC7K1 replication in SAMD9, WDR6, and FTSJ1 CRISPR/Cas9 KO cells. (A) Genome sequences of CRISPR/Cas9 modifications in SAMD9, WDR6, and FTSJ1 genes. The SAMD9, WDR6, and FTSJ1 genes of HeLa cells were modified by Cas9 using guide RNAs that targeted their first exons. Representative cell clones that exhibited enhanced replication of ΔC7K1 were expanded and further analyzed by genome sequencing. Primers that anneal to DNA flanking the Cas9 guide RNA target sites were used for PCR amplification. The PCR products were cloned in plasmids and subjected to Sanger sequencing. Unmodified and CRISPR/Cas9-modified sequences are shown with amino acid codons below the nucleotide sequences. Dashes and asterisks indicate deleted nucleotides and stop codons, respectively. (B) Infection and spread of the C7/K1 deletion mutant. HeLa, SAMD9−/−, WDR6−/−, and FTSJ1−/− cells were infected in triplicate at a multiplicity of infection of 0.03 PFU/cell with ΔC7K1 encoding GFP regulated by the P11 late promoter. After 18 h, GFP-positive cells were counted by flow cytometry. (C) HeLa and SAMD9−/− cells were infected with WT or ΔC7K1 VACV and analyzed as described above. Error bars represent standard errors of the means (SEM). ****, P < 0.0001; **, P < 0.004; *, P < 0.025.