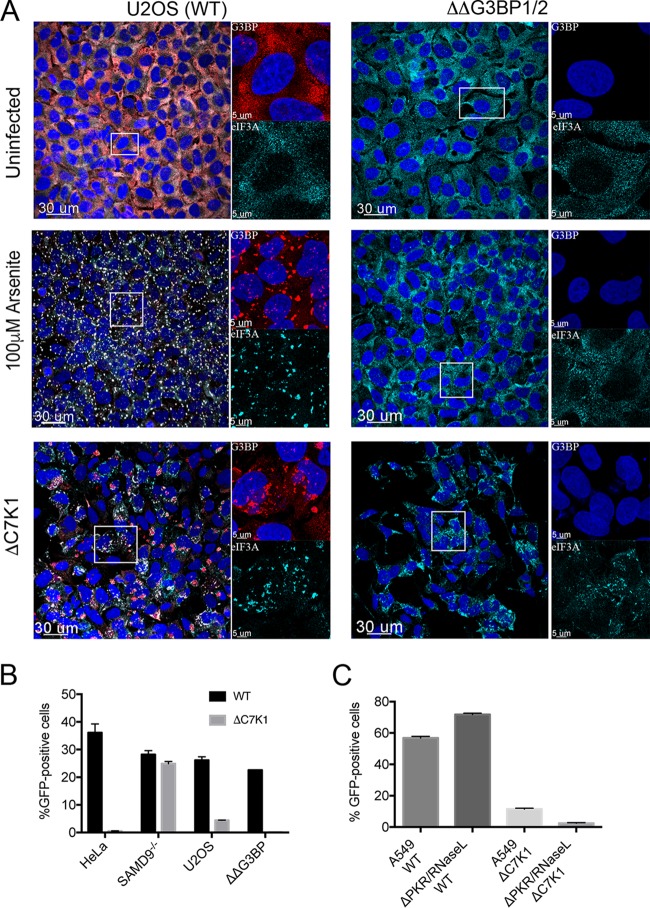
FIG 8.
Effect of G3BP1/2 deletion on granule formation and virus spread. (A) Effect of G3BP1/2 deletion on granule formation. U2OS cells (left) and ΔΔG3BP1/2 U2OS cells (right) were grown on coverslips and left untreated or incubated for 1 h with 100 µM arsenite to induce stress granule formation or inoculated with ΔC7K1 virus at a multiplicity of infection of 3 PFU/cell for 6 h. Cells were fixed and granules were detected with antibodies to G3BP and eIF3A and fluorescent secondary antibodies. Large images on the left represent a merge of DAPI and antibody staining; areas within the boxes are shown at higher magnification on the right as a composite of DAPI and anti-G3BP (upper) and anti-eIF3A (lower). Magnification is indicated by size bars. (B) Effect of G3BP1/2 deletion on spread of ΔC7K1. HeLa, SAMD9−/−, U2OS, and ΔΔG3BP1/2 cells were infected in triplicate with ΔC7K1 at a multiplicity of 0.05 PFU/cell for 18 h. GFP-positive cells were determined by flow cytometry. (C) Effect of PKR and RNase L deletion on spread of ΔC7K1. The procedure was similar to that described for panel B, except that A549 and PKR/RNase L knockout cells were used. Error bars represent SEM.