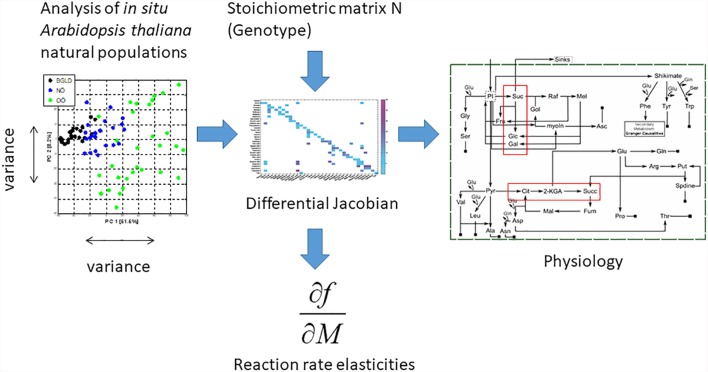
FIGURE 1.
By the application of metabolomics, multivariate statistics and mathematical modeling based on genome-derived biochemical pathway information, biochemical and physiological signatures of in situ Arabidopsis populations can be identified. Different metabolic steady states on a population level and general patterns common to all populations can be distinguished by this metabolic modeling approach in combination with eco-metabolomics. This finally allows for the prediction of characteristic processes of in situ metabolic adaptation. For details see text and Materials and Methods.