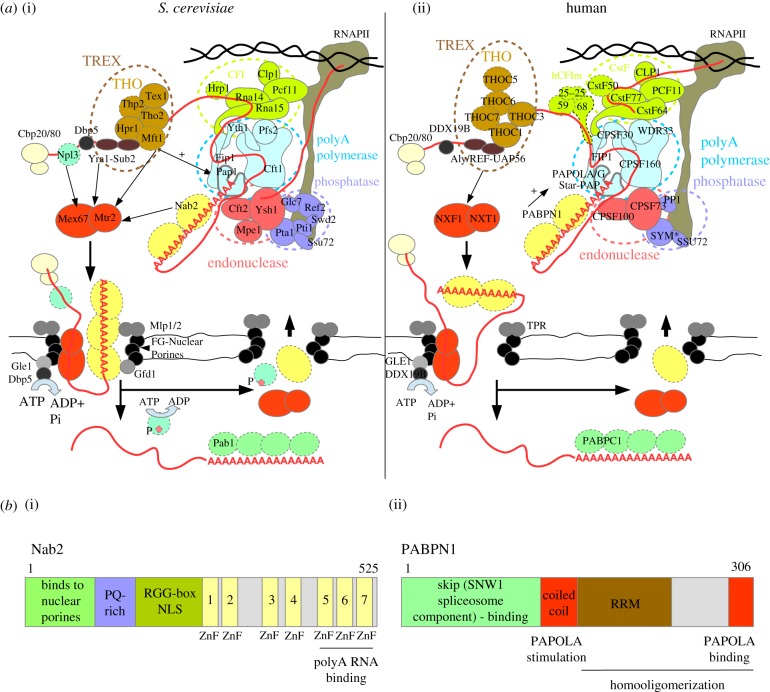
Figure 1.
Model of CPF/CPSF-mediated mRNA 3'-end formation and nuclear export. (a) Schematic overview of 3′-end cleavage, polyadenylation and export factors from S. cerevisiae (a(i)) and human (a(ii)) cells. Factors with no human or S. cerevsiae orthologues with a similar function in polyadenylation and export, respectively, are marked with dashed outlines. In some cases, the human orthologue has not yet been studied in the context of cleavage and polyadenylation. The CPF and CPSF complexes are composed of three basic modules (inspired from model in [20]): (i) the endonuclease module organized around the Ysh1/CPSF73 enzyme supported by Cft2/CPSF100 and Mpe1; (ii) the RNAPII phosphatase module, the catalytic activity of which is mediated by the Glc7/PP1 and Ssu72 subunits of the ‘associated with Pta1′ (human symplekin protein – SYM*) complex (APT: Pta1, Swd2, Ref2, Pti1, Syc1); and (iii) the pA polymerase module, containing the Pap1/PAPOLA/PAPOLG/Star-PAP enzymes, which are recruited to this module mainly by an interaction with Cft1/CPSF160 but also with Fip1/FIP1, Pfs2/WDR33 and Yth1/CPSF30. The yeast CPF complex is further assisted by Cleavage Factor I (CFI: Clp1, Hrp1, Pcf11, Rna14, Rna15), while the human CPSF complex is supported by the CFI-like sub-complexes: cleavage stimulation factor (CstF: CstF50/77/64), cleavage factor II (CFIIm: CLP1, PCF11) and the human-specific cleavage factor I (CFIm: CFIm25/59/68). The CPF and CPSF complexes are recruited dually by binding to specific sequences in the nascent RNA and to the RNAPII CTD. In S. cerevisiae cells, polyadenylation is shown to be stimulated by the THO complex (indicated by the arrow) and in human cells by PABPN1 (arrow). The formation of an export-competent mRNP starts co-transcriptionally with association of the cap-binding complex (CBC, CBP20/80). In yeast Npl3, TREX and Nab2 loading enhances recruitment of the export adaptor Mex67/Mtr2, while in human only the TREX subunit Aly/REF stimulates nuclear export. Successful synthesis of a long pA tail and mRNP assembly leads to release of the newly made RNA from the site of transcription and its translocation to the nuclear pore. Mex67/NXF1 interaction with core FG-nuclear pore factors and Nab2 interactions with the Gfd1 NPC subunit stimulates export. Nab2 binding to Mlp1/Mlp2 promotes retention of unspliced transcripts. An mRNP associated helicase Dbp5/DDX19B is activated by interaction with the Gle1/GLE1 NPC subunit located at the cytoplasmic face and contributes to the displacement of Nab2 and possibly other nuclear mRNP subunits from the transcript. Other mechanisms also contribute to recycling of export factors, such as phosphorylation in case of Npl3. This allows for the release of the transcript into the cytoplasm. Human PABPN1 shuttles between the nucleus and the cytoplasm, but the mechanism by which it is exchanged by cytoplasmic PABPC1 is still unclear. (b) Schematic overview of domain and motif organizations of the main CPF-associated non-orthologous PABs in S. cerevisiae (b(i) Nab2) and human (b(ii) PABPN1) cells highlights the diverse functions of these proteins in pA tail-length control and RNA nuclear export. PABPN1 binds RNA using an RNA Recognition Motif (RRM), while Nab2 recognizes RNA via its three distal zinc fingers (ZnFs) [21]. PABPN1 interacts directly with and stimulates PAPOLA activity, while Nab2 binds the NPC subunits Mlp1/2 and Gfd1 through its N-terminal domain. Moreover, PABPN1 interacts with the spliceosome subunit SKIP [22]. The region responsible for nuclear import of Nab2 is located within the RGG domain [23].