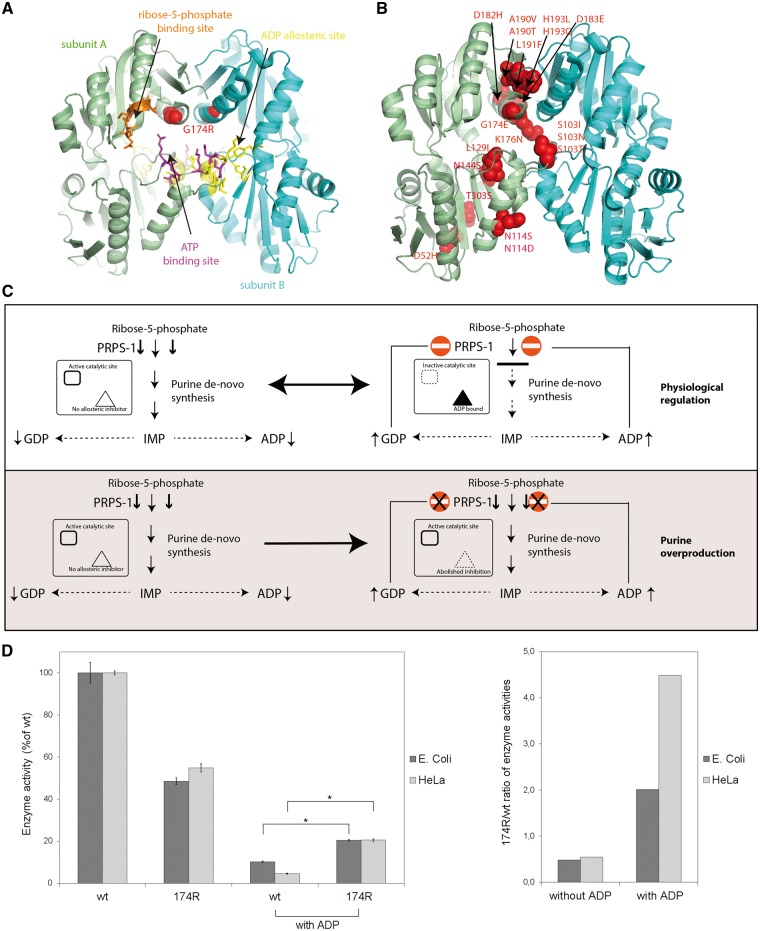
Fig. 2.
Structural and functional studies
(A) Structural mapping of the G174R mutation (red spheres) shows its localization close to PRPS1 binding sites. The ATP binding site (magenta) forms a catalytic pocket jointly with the ribose-5-phosphate site (orange). The ADP allosteric site (yellow) binds allosteric inhibitors of PRPS1. Two subunits of the PRPS1 hexamer are highlighted in a different color. Coordinates of PDB entry 1H06 are used for illustration. (B) Localization of all described PRPS1 mutations causing enzyme superactivity, with most mutations clustered at the dimeric interface. (C) Molecular mechanism of PRPS1 superactive mutations. The mutant enzyme has abolished feedback regulation inducing constitutive activity and purine overproduction. (D) Enzymatic activities of affinity purified recombinant wild-type (wt) PRPS1 and PRPS1 p.174R proteins expressed in E. coli and HeLa cells. Under standard conditions, the PRPS1 p.174R had reduced activity compared with the wt PRPS1. Addition of ADP reduced the activity of the wt PRPS1 much more than the p.174R mutant. Affected feedback regulation of the PRPS1 activity by ADP was demonstrated by increased enzyme activity ratios of PRPS1 p.174R compared with wt PRPS1. Enzyme activities were normalized to that of the wt PRPS1and displayed as mean values of three independent expression experiments; *P < 0.05.