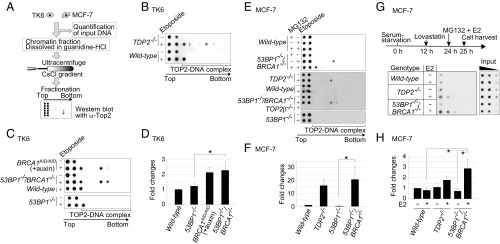
Fig. 3.
Accumulation of TOP2ccs in BRCA1-deficient cells. (A) Schematic of in vivo TOP2cc measurement by immunodetection with α-TOP2 antibody. Genomic DNA (50 μg) (from TK6 and MCF-7) was subjected to sedimentation by CsCl-gradient ultracentrifugation. Genomic DNA from wild-type TK6 cells treated with etoposide (10 μM) for 2 h was included as a control for every dot blot of TK6 cells. The treatment reduced cellular survival by only ∼1% relative to untreated wild-type TK6 cells. Individual fractions were blotted to PVDF filters followed by dot blot using α-TOP2 antibody. The top two fractions include free TOP2 while the bottom fractions include TOP2ccs. (B and C) Dot blot of TOP2 in the indicated TK6 cells treated with etoposide (10 μM) (+) or DMSO (−) for 2 h. BRCA1AID/AID cells were pretreated with auxin for 2 h, and then incubated with etoposide (10 μM) plus auxin for an additional 2 h. (D) Quantification of TOP2cc in the indicated genotypes from C relative to the amount of TOP2cc in wild-type TK6 cells treated with etoposide (10 μM) for 2 h. Each experiment was done independently at least three times. Error bars represent SD. The asterisk indicates P < 0.01, calculated by Student’s t test. (E) TOP2βcc detection in MCF-7 cells. The indicated cells were serum-starved for 24 h and then treated with etoposide (10 μM) for 2 h. The addition of a proteasome inhibitor (MG132) with etoposide caused a signal shift from the middle fractions to the upper-third fraction (Upper). (F) Quantification of etoposide-induced TOP2cc for the indicated genotypes from E, relative to the amount of TOP2cc in wild-type MCF-7. Data are presented as in D. The asterisk indicates P < 0.01, calculated by Student’s t test. (G) Detection of TOP2βcc accumulation by E2 in MCF-7. The diagram (Upper) indicates the experimental design. After incubating MCF-7 cells carrying the indicated genotypes in serum-free medium for 12 h, we added a CDK inhibitor, lovastatin (10 μM), to completely eliminate any cycling cells. We added MG132 and E2 at 24 h after serum starvation and harvested cells at 25 h. MG132 prevents the proteasome degradation of TOP2 at TOP2ccs. Genomic DNA was sheared by sonication. We then conducted immunoprecipitation with α-TOP2β to enrich the TOP2βccs and subjected them to sedimentation by CsCl-gradient ultracentrifugation, as in A. The bottom fractions including TOP2βccs were analyzed by dot blot with α-TOP2β antibody. Threefold serial dilutions of cell lysates were subjected to dot-blot analysis with α–β-actin antibody. (H) Quantification of immunoprecipitated TOP2βccs for the indicated genotypes from G, relative to the amount of immunoprecipitated TOP2cc in wild-type MCF-7 without E2 following normalization with α–β-actin of whole-cell extract (input) as an internal control. Error bars represent SD of three independent experiments. The asterisks indicate P < 0.05, calculated by Student’s t test.